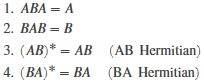【MRI】SENSE (Sensitivity Encoding) 算法 仿真实验与原理剖析 (Matlab 实现) |
您所在的位置:网站首页 › 灵敏度原理 › 【MRI】SENSE (Sensitivity Encoding) 算法 仿真实验与原理剖析 (Matlab 实现) |
【MRI】SENSE (Sensitivity Encoding) 算法 仿真实验与原理剖析 (Matlab 实现)
|
目录 1. 加载全采样 MR 及其敏感度图并显示 1.1 预扫描数据显示 1.2 根据预扫描数据初次生成敏感度图并显示 2. 获取部分参数 3. 将全采样 MR 转换为全采样 k 空间 4. 显示各线圈及 RSOS 组合重建的总体全采样 k 空间频谱 4.1 正常步骤 4.2 根据预扫描数据的 IFFT 再次生成敏感度图并显示 5. 设置加速因子 6. 构造欠采样等距掩模 (equispaced mask) 并显示 7. 模拟/生成欠采样 k 空间数据并显示 8. 欠采样 k 空间数据 IFFT 转换到图像域并显示 9. SENSE 重建 9.1 原理 9.2 实现 9.3 可视化 功能函数 rsos.m (root-sum-of-square) function image = rsos(data) % RSOS Root Sum of Squares Function. % % The root Sum of Squares function necessary for converting 3D multichannel % complex data into 2D real valued data. assert(length(size(data)) == 2 || length(size(data)) == 3, 'Data must be either 2D or 3D.'); image = abs(data) .^ 2; image = sum(image, 3); image = sqrt(image); assert(length(size(image)) == 2); end %% Assignment 2-1: SENSE Reconstruction %% SENSE % (SENSitivity Encoding) parallel imaging reconstruction method. % % See for a basic introduction. % See (p.226) for a more detailed explanation. % % The notation used in the code below is based on the review paper.约定:相位编码方向 k_y 自底向上 (↑)、读出方向/频率编码方向 k_x 从左往右 (→)。 1. 加载全采样 MR 及其敏感度图并显示 1.1 预扫描数据显示brainCoils 是 5 通道的大脑 MR 图像,分别来自 Nc = 5 个线圈各自的全采样结果,每张图像尺寸为 120 ×128。 coilMap 是 5 通道的线圈敏感度图,与 brainCoils 的 shape 或 size 相同,元素值一一对应。该敏感度图来自 预扫描 过程,(本实现暂未涉及),默认正常工作时每次扫描环境下敏感度图相同 (本实现仅用于展示说明,降低严谨性是可容忍的)。 %% Loading data brainCoilsData = load('brain_coil.mat'); brainCoils = brainCoilsData.brain_coil_tmp; coilMapData = load('coil_sensitivity_map.mat'); coilMap = coilMapData.coil_map; % 120×128×5 double接着,分别显示各线圈的全采样 MR 图 brainCoils(:, :, Nc): %% Examine full sampled MR images of each coil figure; colormap('gray'); subplot(2, 3, 1); imagesc(brainCoils(:,:,1)); title('coil-1 full sampled MR image'); colorbar(); subplot(2, 3, 2); imagesc(brainCoils(:,:,2)); title('coil-2 full sampled MR image'); colorbar(); subplot(2, 3, 3); imagesc(brainCoils(:,:,3)); title('coil-3 full sampled MR image'); colorbar(); subplot(2, 3, 4); imagesc(brainCoils(:,:,4)); title('coil-4 full sampled MR image'); colorbar(); subplot(2, 3, 5); imagesc(brainCoils(:,:,5)); title('coil-5 full sampled MR image'); colorbar();
然后,分别显示各线圈的敏感度图 coilMap(:, :, Nc): %% Examine sensitivity map of each coil figure; colormap('gray'); subplot(2, 3, 1); imagesc(coilMap(:,:,1)); title('coil-1 sensitivity map'); colorbar(); subplot(2, 3, 2); imagesc(coilMap(:,:,2)); title('coil-2 sensitivity map'); colorbar(); subplot(2, 3, 3); imagesc(coilMap(:,:,3)); title('coil-3 sensitivity map'); colorbar(); subplot(2, 3, 4); imagesc(coilMap(:,:,4)); title('coil-4 sensitivity map'); colorbar(); subplot(2, 3, 5); imagesc(coilMap(:,:,5)); title('coil-5 sensitivity map'); colorbar();
对比可见,根据公式手动生成的敏感度图不仅具有一些噪点瑕疵,而且对比度也存在差异。虽然 1.1 中预扫描给出的敏感度图可能是经过后处理的,但根本原因在于这种计算方式不符合实际,毕竟 MRI 系统实际先采集到的应该是 raw k-space 而非 MR image。为此,真正的计算方式详见 4.2 节。 敏感度图的计算 计算线圈敏感度是 SENSE 过程中初始而最重要的一步。低分辨率图像是在 全 FOV 下从各表面线圈分别采集的。这些表面线圈图像通过将其除以低分辨率总体线圈图像进行归一化。然后对数据进行 滤波、阈值化和点估计,以生成线圈敏感度图(如右图所示)。这些映射量化每个线圈接收区域内不同原点的信号的相对权重。
一旦线圈敏感度图被计算出来,正式工作的磁共振脉冲序列就开始了。 注意,SENSE 假定后续每次扫描时,敏感度图都是不变的。然而,事实上每次扫描的系统环境可能都有所不同,敏感度图会随扫描次数而发生 (细节性的) 变化。 2. 获取部分参数phaseLength 表示 k 空间相位编码方向长度 ky_FOV = 120。(图像域 MR 图像/视野高度 FOV_y = 120) freqLength 表示 k 空间频率编码方向长度 kx_FOV = 128。(图像域 MR 图像/视野宽度 FOV_x = 128) coilNum 表示采用线圈数 Nc = 5。 %% size [phaseLength, freqLength, coilNum] = size(brainCoils); % ky_FOV=120, kx_FOV=128, Nc=5 disp(size(brainCoils)); % Phases, Frequencies, Coil Number 3. 将全采样 MR 转换为全采样 k 空间fullKpace 为各线圈全采样 MR 图像对应的 k 空间域数据,size = 120×128×5,type = complex double %% Convert to Fourier Domain with 2D Fast Fourier Transform % Remember to use fftshift and ifftshift when using the Fourier Transform. % Otherwise, the k-space will not be centered properly. fullKspace = ifftshift(fft2(fftshift(brainCoils))); % 注意转换到 k 空间时, FFT 前后要分别 shiftfftshift 与 ifftshift 注意,当空域中的点 f(x,y) 移位时,在频域中只发生相移,并不影响其傅里叶变换的幅度。反之,当频域中的点 F(u,v) 移位时,相应的 f(x,y) 在空域中也只发生相移,不产生幅值变化。根据平移性质,为更清楚查看频率域的 k 空间数据 的频谱 (复数值数据的模),在把画面分成四分的基础上,可以进行换位/移位 (shift),从而使频域原点 (直流成分) 平移到频谱中央。如下所示:
举个例子:
fullKspaceImage 是通过 平方和根 (root-sum-of-square) 方法 组合 所有线圈各自的全采样 k 空间 复数值数据 重建的 总体全采样 k 空间 实数值频谱,size = 120×128,type = double rsos(fullKspace(:, :, Nc) 则分别为由 各线圈各自的全采样 k 空间 复数值数据 取模得到的 实数值频谱,size = 120×128,type = double %% Examine raw k-space. fullKspaceImage = rsos(fullKspace); % RSOS 重建得到实值全采样 k 空间频谱图像 figure; colormap('gray'); subplot(2, 3, 1); imagesc(rsos(fullKspace(:,:,1)), [0, 10000]); title('coil-1 k-space'); colorbar(); subplot(2, 3, 2); imagesc(rsos(fullKspace(:,:,2)), [0, 10000]); title('coil-2 k-space'); colorbar(); subplot(2, 3, 3); imagesc(rsos(fullKspace(:,:,3)), [0, 10000]); title('coil-3 k-space'); colorbar(); subplot(2, 3, 4); imagesc(rsos(fullKspace(:,:,4)), [0, 10000]); title('coil-4 k-space'); colorbar(); subplot(2, 3, 5); imagesc(rsos(fullKspace(:,:,5)), [0, 10000]); title('coil-5 k-space'); colorbar(); subplot(2, 3, 6); imagesc(fullKspaceImage, [0, 10000]); title('RSOS reconstruction k-space'); colorbar();
事实上,MRI 系统采集到的本应就是 raw k-space 数据,只不过代码给的测试文件是图像域 MR 图像而已。 %% =================================== 专门测试敏感度图 ===================================== rawfullKspace = fullKspace; acsBrainCoils = ifftshift(ifft2(fftshift(rawfullKspace))); % 128×120×5 complex double acsImage = rsos(acsBrainCoils); % RSOS 重建 128×120 double figure; colormap('gray'); subplot(2, 3, 1); sens_map_1 = rsos(acsBrainCoils(:,:,1)) ./ acsImage; imagesc(sens_map_1); title('coil-1 sensitivity map (manual)'); colorbar(); subplot(2, 3, 2); sens_map_2 = rsos(acsBrainCoils(:,:,2)) ./ acsImage; imagesc(sens_map_2); title('coil-2 sensitivity map (manual)'); colorbar(); subplot(2, 3, 3); sens_map_3 = rsos(acsBrainCoils(:,:,3)) ./ acsImage; imagesc(sens_map_3); title('coil-3 sensitivity map (manual)'); colorbar(); subplot(2, 3, 4); sens_map_4 = rsos(acsBrainCoils(:,:,4)) ./ acsImage; imagesc(sens_map_4); title('coil-4 sensitivity map (manual)'); colorbar(); subplot(2, 3, 5); sens_map_5 = rsos(acsBrainCoils(:,:,5)) ./ acsImage; imagesc(sens_map_5); title('coil-5 sensitivity map (manual)'); colorbar(); subplot(2, 3, 6); imagesc(acsImage); title('RSOS reconstruction body MR image'); colorbar();
可以看到,本次生成的敏感度图与程序初始输入的敏感度图十分接近了! 5. 设置加速因子downSamplingRate 为下/欠采样率,又称缩减因子 (reduction factor) / 加速因子 (accelation factor),符号通常为 R。此处设置 R = 5 表示将进行 5 倍加速 MRI,这在笛卡尔 k 空间顺序采集中,意味着每 R = 5 条相位编码线的位置才真正地采集到 1 条 (acquired line),其位置将由 1 值 mask 覆盖表示已采集;其余 4 条 (unacquired line) 的位置将由 0 值 mask 覆盖表示未采集。从而,实现 R = 5 倍欠采样 / 加速。 %% setting reduction/accelation factor downSamplingRate = 5; % 缩减/加速因子 R = 5 —— 5 倍欠采样/加速 6. 构造欠采样等距掩模 (equispaced mask) 并显示fullKpace 为各线圈全采样 MR 图像对应的 k 空间域数据,size = 120×128×5,type = complex double mask 为与 fullKpace 等 size 和 type 的 0/1 等距掩模 (不是随机掩模),用于模拟/制作 R = 5 倍欠采样 k 空间数据。 %% Making the mask % Creating a mask for the downsampling and the ACS lines. mask = zeros(size(fullKspace)); % Making a mask for the brain coils. for line = 1:phaseLength % Goes along the phase encoding axis. (vertical direction ky - dim=0) if rem(line, downSamplingRate) == 0 % r = rem(a,b) returns the remainder after division of a by b, mask(line, :, :) = 1; % Broadcasting the value of 1 to mask(i, :, :). % 每 R 行采集一条相位编码线, mask=1 end end assert(isequal(size(mask), size(brainCoils)), 'Mask size is incorrect.') % mask 要和 full FOV MR image 等尺寸然后,显示构造好的 R = 5 倍欠采样等距 mask: %% Displaying mask. % This code functions to check whether the mask has been made correctly. % White lines indicate 1's. Black lines indicate 0's. All values should be either 0 or 1. % There should be a white band in the middle, with striped lines surrounding it. % 取均值其实没什么特殊意义, 只是把 5 线圈对应的 5 通道压缩成单通道而已 % dispMask = mask(:, :, 1); % 用 mask(:,:,1) 单独取出一个通道也行, 2,3,4,5 均亦可 dispMask = mean(mask, 3); % M = mean(A,dim) returns the mean along dimension dim figure; imagesc(dispMask); colormap('gray'); colorbar(); title('equispaced mask (R = 5)')
fullKpace 为各线圈全采样 MR 图像对应的 k 空间域数据,size = 120×128×5,type = complex double mask 为 0/1 等距掩模,用于模拟/生成 R = 5 倍欠采样 k 空间数据,size = 120×128×5,type = complex double downSampledKpace 为 fullKpace 和 mask 通过阵列乘法 —— 按元素相乘得到的模拟 R = 5 倍欠采样 k 空间复数值数据,size = 120×128×5,type = complex double %% Generating the down-sampled k-space image % Obtain the Hadamard product (elementwise matrix multiplication) between the % full k-space data and the mask. % % Hint: use the .* operator, not the * operator, which does matrix multiplication. % Elementwise (Hadamard) product (multiplication) of matrices. % 阵列乘法 —— 按元素相乘 % 5 个 fullKspace 各不相同 (位置及敏感度差异导致的观测区别) downSampledKspace = fullKspace .* mask; % 用 masked data 模拟欠采样 k-space (复数值) 128×120×5 complex doubledownSampleKspaceImage 是通过 平方和根 (root-sum-of-square) 方法 组合 所有线圈各自的欠采样 masked k 空间 复数值数据 重建的 总体欠采样 masked k 空间 实数值频谱,size = 120×128,type = double downSampleKspaceImage_Nc 则分别为由 各线圈各自的欠采样 masked k 空间 复数值数据 取模得到的 实数值频谱,size = 120×128,type = double %% Confirmation % Confirming that the masking has been performed properly in k-space. % 实数值频谱, 仅用于展示, 无实际用途 downSampledKspaceImage = rsos(downSampledKspace); % RSOS 组合多个欠采样 k 空间, 重建得到 实值欠采样 k 空间频谱图像 (多对一) 128×120 double % figure; % imagesc(downSampledKspaceImage, [0, 10000]); % colormap('gray'); % colorbar(); assert(isequal(size(downSampledKspace), size(brainCoils)), 'Reconstruction is of the wrong shape.') figure; colormap('gray'); subplot(2, 3, 1); downSampledKspaceImage_1 = rsos(downSampledKspace(:,:,1)); imagesc(downSampledKspaceImage_1, [0, 10000]); title('coil-1 masked k-space'); colorbar(); subplot(2, 3, 2); downSampledKspaceImage_2 = rsos(downSampledKspace(:,:,2)); imagesc(downSampledKspaceImage_2, [0, 10000]); title('coil-2 masked k-space'); colorbar(); subplot(2, 3, 3); downSampledKspaceImage_3 = rsos(downSampledKspace(:,:,3)); imagesc(downSampledKspaceImage_3, [0, 10000]); title('coil-3 masked k-space'); colorbar(); subplot(2, 3, 4); downSampledKspaceImage_4 = rsos(downSampledKspace(:,:,4)); imagesc(downSampledKspaceImage_4, [0, 10000]); title('coil-4 masked k-space'); colorbar(); subplot(2, 3, 5); downSampledKspaceImage_5 = rsos(downSampledKspace(:,:,5)); imagesc(downSampledKspaceImage_5, [0, 10000]); title('coil-5 masked k-space'); colorbar(); subplot(2, 3, 6); imagesc(downSampledKspaceImage, [0, 10000]); title('RSOS reconstruction masked k-space'); colorbar();
dsBrainCoils 为 R = 5 倍欠采样 k 空间 复数值数据 的 downSampleKspace 图像域 MR 图像 (Nc = 5 个线圈各自的都叠在一起),size = 120×128×5,type = complex double %% Returning the downsampled data to image space. %% complex sub-sampled MR image dsBrainCoils = ifftshift(ifft2(fftshift(downSampledKspace))); % 注意转换到图像域时, IFFT 前后要分别 shift 120x128x5 complex double assert(isequal(size(dsBrainCoils), size(brainCoils)), 'Image shape is wrong.');dsImage 是通过 平方和根 (root-sum-of-square) 方法 组合 所有线圈各自的欠采样 MR 复数值图像数据 dsBrainCoils 而重建的 总体欠采样 MR 实数值图像,size = 120×128,type = double dsImage_Nc 则分别为由 各线圈各自的欠采样 MR 复数值图像数据 取模得到的 实数值 MR 图像,size = 120×128,type = double %% Assignment 2-1: SENSE Reconstruction %% SENSE % (SENSitivity Encoding) parallel imaging reconstruction method. % % See for a basic introduction. % See (p.226) for a more detailed explanation. % % The notation used in the code below is based on the review paper. %% Loading data brainCoilsData = load('brain_coil.mat'); brainCoils = brainCoilsData.brain_coil_tmp; % 120×128×5 double coilMapData = load('coil_sensitivity_map.mat'); coilMap = coilMapData.coil_map; % 120×128×5 double %% Examine full sampled MR images of each coil figure; colormap('gray'); subplot(2, 3, 1); imagesc(brainCoils(:,:,1)); title('coil-1 full sampled MR image'); colorbar(); subplot(2, 3, 2); imagesc(brainCoils(:,:,2)); title('coil-2 full sampled MR image'); colorbar(); subplot(2, 3, 3); imagesc(brainCoils(:,:,3)); title('coil-3 full sampled MR image'); colorbar(); subplot(2, 3, 4); imagesc(brainCoils(:,:,4)); title('coil-4 full sampled MR image'); colorbar(); subplot(2, 3, 5); imagesc(brainCoils(:,:,5)); title('coil-5 full sampled MR image'); colorbar(); %% Examine sensitivity map of each coil figure; colormap('gray'); subplot(2, 3, 1); imagesc(coilMap(:,:,1)); title('coil-1 sensitivity map'); colorbar(); subplot(2, 3, 2); imagesc(coilMap(:,:,2)); title('coil-2 sensitivity map'); colorbar(); subplot(2, 3, 3); imagesc(coilMap(:,:,3)); title('coil-3 sensitivity map'); colorbar(); subplot(2, 3, 4); imagesc(coilMap(:,:,4)); title('coil-4 sensitivity map'); colorbar(); subplot(2, 3, 5); imagesc(coilMap(:,:,5)); title('coil-5 sensitivity map'); colorbar(); %% test about generating sensitivity map manually BodybrainCoils = rsos(brainCoils); % RSOS reconstruction for body MR image figure; colormap('gray'); subplot(2, 3, 1); sensitivity_map_1 = brainCoils(:,:,1) ./ BodybrainCoils; imagesc(sensitivity_map_1); title('coil-1 sensitivity map (manual)'); colorbar(); subplot(2, 3, 2); sensitivity_map_2 = brainCoils(:,:,2) ./ BodybrainCoils; imagesc(sensitivity_map_2); title('coil-2 sensitivity map (manual)'); colorbar(); subplot(2, 3, 3); sensitivity_map_3 = brainCoils(:,:,3) ./ BodybrainCoils; imagesc(sensitivity_map_3); title('coil-3 sensitivity map (manual)'); colorbar(); subplot(2, 3, 4); sensitivity_map_4 = brainCoils(:,:,4) ./ BodybrainCoils; imagesc(sensitivity_map_4); title('coil-4 sensitivity map (manual)'); colorbar(); subplot(2, 3, 5); sensitivity_map_5 = brainCoils(:,:,5) ./ BodybrainCoils; imagesc(sensitivity_map_5); title('coil-5 sensitivity map (manual)'); colorbar(); subplot(2, 3, 6); imagesc(BodybrainCoils); title('RSOS reconstruction body MR image'); colorbar(); %% size [phaseLength, freqLength, coilNum] = size(brainCoils); % 120, 128, Nc=5 disp(size(brainCoils)); % Phases, Frequencies, Coil Number %% Convert to Fourier Domain with 2D Fast Fourier Transform % Remember to use fftshift and ifftshift when using the Fourier Transform. % Otherwise, the k-space will not be centered properly. fullKspace = ifftshift(fft2(fftshift(brainCoils))); % 注意转换到 k 空间时, FFT 前后要分别 shift 120×128×5 complex double %% Examine raw k-space. fullKspaceImage = rsos(fullKspace); % RSOS 重建得到实值全采样 k 空间频谱图像 figure; colormap('gray'); subplot(2, 3, 1); imagesc(rsos(fullKspace(:,:,1)), [0, 10000]); title('coil-1 k-space'); colorbar(); subplot(2, 3, 2); imagesc(rsos(fullKspace(:,:,2)), [0, 10000]); title('coil-2 k-space'); colorbar(); subplot(2, 3, 3); imagesc(rsos(fullKspace(:,:,3)), [0, 10000]); title('coil-3 k-space'); colorbar(); subplot(2, 3, 4); imagesc(rsos(fullKspace(:,:,4)), [0, 10000]); title('coil-4 k-space'); colorbar(); subplot(2, 3, 5); imagesc(rsos(fullKspace(:,:,5)), [0, 10000]); title('coil-5 k-space'); colorbar(); subplot(2, 3, 6); imagesc(fullKspaceImage, [0, 10000]); title('RSOS reconstruction k-space'); colorbar(); %% Setting parameters for later use. %% setting reduction/accelation factor downSamplingRate = 5; % 缩减/加速因子 R = 5 —— 5 倍欠采样/加速 %% Making the mask % Creating a mask for the downsampling and the ACS lines. mask = zeros(size(fullKspace)); % Making a mask for the brain coils. for line = 1:phaseLength % Goes along the phase encoding axis. (vertical direction ky - dim=0) if rem(line, downSamplingRate) == 0 % r = rem(a,b) returns the remainder after division of a by b, mask(line, :, :) = 1; % Broadcasting the value of 1 to mask(i, :, :). % 每 R 行采集一条相位编码线, mask=1 end end assert(isequal(size(mask), size(brainCoils)), 'Mask size is incorrect.') % mask 要和 full FOV MR image 等尺寸 %% Displaying mask. % This code functions to check whether the mask has been made correctly. % White lines indicate 1's. Black lines indicate 0's. All values should be either 0 or 1. % There should be a white band in the middle, with striped lines surrounding it. % 取均值其实没什么特殊意义, 只是把 5 线圈对应的 5 通道压缩成单通道而已 % dispMask = mask(:, :, 1); % 用 mask(:,:,1) 单独取出一个通道也行, 2,3,4,5 均亦可 dispMask = mean(mask, 3); % M = mean(A,dim) returns the mean along dimension dim figure; imagesc(dispMask); colormap('gray'); colorbar(); title('equispaced mask (R = 5)') %% Generating the down-sampled k-space image % Obtain the Hadamard product (elementwise matrix multiplication) between the % full k-space data and the mask. % % Hint: use the .* operator, not the * operator, which does matrix multiplication. % Elementwise (Hadamard) product (multiplication) of matrices. % 阵列乘法 —— 按元素相乘 % 5 个 fullKspace 各不相同 (位置及敏感度差异导致的观测区别) downSampledKspace = fullKspace .* mask; % 用 masked data 模拟欠采样 k-space (复数值) 128×120×5 complex double %% Confirmation % Confirming that the masking has been performed properly in k-space. % 实数值频谱, 仅用于展示, 无实际用途 downSampledKspaceImage = rsos(downSampledKspace); % RSOS 组合多个欠采样 k 空间, 重建得到 实值欠采样 k 空间频谱图像 (多对一) 128×120 double % figure; % imagesc(downSampledKspaceImage, [0, 10000]); % colormap('gray'); % colorbar(); assert(isequal(size(downSampledKspace), size(brainCoils)), 'Reconstruction is of the wrong shape.') figure; colormap('gray'); subplot(2, 3, 1); downSampledKspaceImage_1 = rsos(downSampledKspace(:,:,1)); imagesc(downSampledKspaceImage_1, [0, 10000]); title('coil-1 masked k-space'); colorbar(); subplot(2, 3, 2); downSampledKspaceImage_2 = rsos(downSampledKspace(:,:,2)); imagesc(downSampledKspaceImage_2, [0, 10000]); title('coil-2 masked k-space'); colorbar(); subplot(2, 3, 3); downSampledKspaceImage_3 = rsos(downSampledKspace(:,:,3)); imagesc(downSampledKspaceImage_3, [0, 10000]); title('coil-3 masked k-space'); colorbar(); subplot(2, 3, 4); downSampledKspaceImage_4 = rsos(downSampledKspace(:,:,4)); imagesc(downSampledKspaceImage_4, [0, 10000]); title('coil-4 masked k-space'); colorbar(); subplot(2, 3, 5); downSampledKspaceImage_5 = rsos(downSampledKspace(:,:,5)); imagesc(downSampledKspaceImage_5, [0, 10000]); title('coil-5 masked k-space'); colorbar(); subplot(2, 3, 6); imagesc(downSampledKspaceImage, [0, 10000]); title('RSOS reconstruction masked k-space'); colorbar(); %% Returning the downsampled data to image space. %% complex sub-sampled MR image dsBrainCoils = ifftshift(ifft2(fftshift(downSampledKspace))); % 注意转换到图像域时, IFFT 前后要分别 shift 120x128x5 complex double assert(isequal(size(dsBrainCoils), size(brainCoils)), 'Image shape is wrong.'); %% Checking reconstructed image. %% % Using rsos (root sum of squares) to make visualization possible. dsImage = rsos(dsBrainCoils); % RSOS 重建得到 实值欠采样空间域混叠 MR 图像 120×128 double % figure; % imagesc(dsImage); % colormap('gray'); % colorbar(); figure; colormap('gray'); subplot(2, 3, 1); dsImage_1 = rsos(dsBrainCoils(:,:,1)) ; imagesc(dsImage_1); colormap('gray'); colorbar(); title('coil-1 partial FOV MR image'); subplot(2, 3, 2); dsImage_2 = rsos(dsBrainCoils(:,:,2)) ; imagesc(dsImage_2); colormap('gray'); colorbar(); title('coil-2 partial FOV MR image'); subplot(2, 3, 3); dsImage_3 = rsos(dsBrainCoils(:,:,3)) ; imagesc(dsImage_3); colormap('gray'); colorbar(); title('coil-3 partial FOV MR image'); subplot(2, 3, 4); dsImage_4 = rsos(dsBrainCoils(:,:,4)) ; imagesc(dsImage_4); colormap('gray'); colorbar(); title('coil-4 partial FOV MR image'); subplot(2, 3, 5); dsImage_5 = rsos(dsBrainCoils(:,:,5)) ; imagesc(dsImage_5); colormap('gray'); colorbar(); title('coil-5 partial FOV MR image'); subplot(2, 3, 6); imagesc(dsImage); title('RSOS reconstruction partial FOV MR image'); colorbar(); %% SENSE Reconstruction %% % Not a perfect solution but works for this experiment. fieldOfView = floor(phaseLength/downSamplingRate); % FOV_y = 120 / 5 = 24 % FOV_x = 128 senseRecon = zeros(phaseLength, freqLength); % 120×128 complex double - MR 重建结果 vectorI = zeros(coilNum, 1); % 5×1 complex double - 混叠像素向量(5个线圈在同一像素位置的复值) cHat = zeros(coilNum, downSamplingRate); % 5×5 double - 敏感度图(线圈数×重叠像素数) for x = 1:freqLength % kx = FOV_x = 128 for y = 1:fieldOfView % FOV_y = 24 % 欠采样 MR 图像在图像域 pixel(x, y) 处的 Nc=5 个混叠像素值存入 5×1 complex double 向量 I vectorI = reshape(dsBrainCoils(y, x, :), coilNum, 1); % All channels of single pixel in image. % 每个混叠像素 pixel(x, y) 都由 R = 5 个 desired full MR image 的像素重叠而成 for k = 1:downSamplingRate % R = 5 % Coil sensitivities of all channels of a single pixel per segment. cHat(:, k) = reshape(coilMap(y + (k-1) .* fieldOfView, x, :), coilNum, 1); % 5×1 double % y + (k-1).*fieldOfView 是敏感度图 FOV_y 方向上的偏移量 % cHat(:, k) 表示所有 5 个线圈在第 k 个 desired full MR image 像素点处的敏感度值 end % Eq.4 cHatPinv = pinv(cHat); % Moore-Penrose Pseudoinverse. vectorRho = cHatPinv * vectorI; % 5×1 complex double - 5 个 desired full MR image 像素点 for k = 1:downSamplingRate % 按 FOV_y 方向上的偏移量等距保存 desired full MR image 像素点 senseRecon(y + (k-1) .* fieldOfView, x) = vectorRho(k); end end end %% Visualizing results %% senseImage = rsos(senseRecon); senseImage = senseImage .* downSamplingRate; % Necessary to correct for the missing values. original = rsos(brainCoils); delta = original-senseImage; figure; colormap('gray'); subplot(2, 2, 1); imagesc(original); title('Original Image'); colorbar(); subplot(2, 2, 2); imagesc(dsImage); title('Downsampled Image'); colorbar(); subplot(2, 2, 3); imagesc(senseImage); title('SENSE Image'); colorbar(); subplot(2, 2, 4); imagesc(delta); title('Difference Image'); colorbar();
为什么欠采样 k 空间在 ky 方向间隔增大 5 倍,IFFT 到图像域时却非 1/5 FOV_y 的 MR 图像? 注意,根据《数字信号处理》理论,频域中的有限离散值,在时域中对应的有限长序列为:原序列以采样点数为周期进行周期延拓后的主值序列。 因此,此处每张混叠部分 FOV MR 图像显示的是 长度为 120 / 5 = 24 的主值序列的 R = 5 次周期延拓的结果,因此呈现的混叠部分 FOV MR 图像视野高度 FOV_y 仍为 120。若要得到理论上的长度为 24 的混叠部分 FOV MR 图像,只需任取 R = 5 个周期中的任一者即可。例如,对于 RSOS 重建的混叠部分 FOV MR 图像 (最后一张),其理论形象为 (1/5 FOV_y):
Moore-Penrose 伪逆 Moore-Penrose 伪逆 是一种矩阵,可在不存在逆矩阵的情况下作为逆矩阵的部分替代。此矩阵常被用于求解没有唯一解或有许多解的线性方程组。 对于任何矩阵 A 来说,伪逆 B 都存在,是唯一的,并且具有与 A' (A 的转置) 相同的维度。若 A 是方阵且非奇异,则 pinv(A) 只是一种成本比较高的计算 inv(A) 的方式。但若 A 不是方阵,或是方阵且奇异,则 inv(A) 不存在。在这些情况下,pinv(A) 拥有 inv(A) 的部分(但非全部)属性:
伪逆计算基于 svd(A)。该计算将小于 tol 的奇异值视为零。 具体地:
pinv 通过奇异值分解来形成 A 的伪逆。S 对角线上小于奇异值容差 tol 的奇异值 (不那么重要或代表性不够的特征值) 被视为零,而 A 的表示由此变成:
因此 A 的伪逆等于:
参考文献:Moore-Penrose 伪逆 - MATLAB pinv- MathWorks 中国 9.3 可视化 %% Visualizing results %% senseImage = rsos(senseRecon); senseImage = senseImage .* downSamplingRate; % Necessary to correct for the missing values. original = rsos(brainCoils); delta = original-senseImage; figure; colormap('gray'); subplot(2, 2, 1); imagesc(original); title('Original Image'); colorbar(); subplot(2, 2, 2); imagesc(dsImage); title('Downsampled Image'); colorbar(); subplot(2, 2, 3); imagesc(senseImage); title('SENSE Image'); colorbar(); subplot(2, 2, 4); imagesc(delta); title('Difference Image'); colorbar();
完整程序 SENSE.m %% Assignment 2-1: SENSE Reconstruction %% SENSE % (SENSitivity Encoding) parallel imaging reconstruction method. % % See for a basic introduction. % See (p.226) for a more detailed explanation. % % The notation used in the code below is based on the review paper. %% Loading data brainCoilsData = load('brain_coil.mat'); brainCoils = brainCoilsData.brain_coil_tmp; % 120×128×5 double coilMapData = load('coil_sensitivity_map.mat'); coilMap = coilMapData.coil_map; % 120×128×5 double %% Examine full sampled MR images of each coil figure; colormap('gray'); subplot(2, 3, 1); imagesc(brainCoils(:,:,1)); title('coil-1 full sampled MR image'); colorbar(); subplot(2, 3, 2); imagesc(brainCoils(:,:,2)); title('coil-2 full sampled MR image'); colorbar(); subplot(2, 3, 3); imagesc(brainCoils(:,:,3)); title('coil-3 full sampled MR image'); colorbar(); subplot(2, 3, 4); imagesc(brainCoils(:,:,4)); title('coil-4 full sampled MR image'); colorbar(); subplot(2, 3, 5); imagesc(brainCoils(:,:,5)); title('coil-5 full sampled MR image'); colorbar(); %% Examine sensitivity map of each coil figure; colormap('gray'); subplot(2, 3, 1); imagesc(coilMap(:,:,1)); title('coil-1 sensitivity map'); colorbar(); subplot(2, 3, 2); imagesc(coilMap(:,:,2)); title('coil-2 sensitivity map'); colorbar(); subplot(2, 3, 3); imagesc(coilMap(:,:,3)); title('coil-3 sensitivity map'); colorbar(); subplot(2, 3, 4); imagesc(coilMap(:,:,4)); title('coil-4 sensitivity map'); colorbar(); subplot(2, 3, 5); imagesc(coilMap(:,:,5)); title('coil-5 sensitivity map'); colorbar(); %% size [phaseLength, freqLength, coilNum] = size(brainCoils); % 120, 128, Nc=5 disp(size(brainCoils)); % Phases, Frequencies, Coil Number %% Convert to Fourier Domain with 2D Fast Fourier Transform % Remember to use fftshift and ifftshift when using the Fourier Transform. % Otherwise, the k-space will not be centered properly. fullKspace = ifftshift(fft2(fftshift(brainCoils))); % 注意转换到 k 空间时, FFT 前后要分别 shift 120×128×5 complex double %% Examine raw k-space. fullKspaceImage = rsos(fullKspace); % RSOS 重建得到实值全采样 k 空间频谱图像 figure; colormap('gray'); subplot(2, 3, 1); imagesc(rsos(fullKspace(:,:,1)), [0, 10000]); title('coil-1 k-space'); colorbar(); subplot(2, 3, 2); imagesc(rsos(fullKspace(:,:,2)), [0, 10000]); title('coil-2 k-space'); colorbar(); subplot(2, 3, 3); imagesc(rsos(fullKspace(:,:,3)), [0, 10000]); title('coil-3 k-space'); colorbar(); subplot(2, 3, 4); imagesc(rsos(fullKspace(:,:,4)), [0, 10000]); title('coil-4 k-space'); colorbar(); subplot(2, 3, 5); imagesc(rsos(fullKspace(:,:,5)), [0, 10000]); title('coil-5 k-space'); colorbar(); subplot(2, 3, 6); imagesc(fullKspaceImage, [0, 10000]); title('RSOS reconstruction k-space'); colorbar(); %% Setting parameters for later use. %% setting reduction/accelation factor downSamplingRate = 5; % 缩减/加速因子 R = 5 —— 5 倍欠采样/加速 %% Making the mask % Creating a mask for the downsampling and the ACS lines. mask = zeros(size(fullKspace)); % Making a mask for the brain coils. for line = 1:phaseLength % Goes along the phase encoding axis. (vertical direction ky - dim=0) if rem(line, downSamplingRate) == 0 % r = rem(a,b) returns the remainder after division of a by b, mask(line, :, :) = 1; % Broadcasting the value of 1 to mask(i, :, :). % 每 R 行采集一条相位编码线, mask=1 end end assert(isequal(size(mask), size(brainCoils)), 'Mask size is incorrect.') % mask 要和 full FOV MR image 等尺寸 %% Displaying mask. % This code functions to check whether the mask has been made correctly. % White lines indicate 1's. Black lines indicate 0's. All values should be either 0 or 1. % There should be a white band in the middle, with striped lines surrounding it. % 取均值其实没什么特殊意义, 只是把 5 线圈对应的 5 通道压缩成单通道而已 % dispMask = mask(:, :, 1); % 用 mask(:,:,1) 单独取出一个通道也行, 2,3,4,5 均亦可 dispMask = mean(mask, 3); % M = mean(A,dim) returns the mean along dimension dim figure; imagesc(dispMask); colormap('gray'); colorbar(); title('equispaced mask (R = 5)') %% Generating the down-sampled k-space image % Obtain the Hadamard product (elementwise matrix multiplication) between the % full k-space data and the mask. % % Hint: use the .* operator, not the * operator, which does matrix multiplication. % Elementwise (Hadamard) product (multiplication) of matrices. % 阵列乘法 —— 按元素相乘 % 5 个 fullKspace 各不相同 (位置及敏感度差异导致的观测区别) downSampledKspace = fullKspace .* mask; % 用 masked data 模拟欠采样 k-space (复数值) 128×120×5 complex double %% Confirmation % Confirming that the masking has been performed properly in k-space. % 实数值频谱, 仅用于展示, 无实际用途 downSampledKspaceImage = rsos(downSampledKspace); % RSOS 组合多个欠采样 k 空间, 重建得到 实值欠采样 k 空间频谱图像 (多对一) 128×120 double % figure; % imagesc(downSampledKspaceImage, [0, 10000]); % colormap('gray'); % colorbar(); assert(isequal(size(downSampledKspace), size(brainCoils)), 'Reconstruction is of the wrong shape.') figure; colormap('gray'); subplot(2, 3, 1); downSampledKspaceImage_1 = rsos(downSampledKspace(:,:,1)); imagesc(downSampledKspaceImage_1, [0, 10000]); title('coil-1 masked k-space'); colorbar(); subplot(2, 3, 2); downSampledKspaceImage_2 = rsos(downSampledKspace(:,:,2)); imagesc(downSampledKspaceImage_2, [0, 10000]); title('coil-2 masked k-space'); colorbar(); subplot(2, 3, 3); downSampledKspaceImage_3 = rsos(downSampledKspace(:,:,3)); imagesc(downSampledKspaceImage_3, [0, 10000]); title('coil-3 masked k-space'); colorbar(); subplot(2, 3, 4); downSampledKspaceImage_4 = rsos(downSampledKspace(:,:,4)); imagesc(downSampledKspaceImage_4, [0, 10000]); title('coil-4 masked k-space'); colorbar(); subplot(2, 3, 5); downSampledKspaceImage_5 = rsos(downSampledKspace(:,:,5)); imagesc(downSampledKspaceImage_5, [0, 10000]); title('coil-5 masked k-space'); colorbar(); subplot(2, 3, 6); imagesc(downSampledKspaceImage, [0, 10000]); title('RSOS reconstruction masked k-space'); colorbar(); %% Returning the downsampled data to image space. %% complex sub-sampled MR image dsBrainCoils = ifftshift(ifft2(fftshift(downSampledKspace))); % 注意转换到图像域时, IFFT 前后要分别 shift 120x128x5 complex double assert(isequal(size(dsBrainCoils), size(brainCoils)), 'Image shape is wrong.'); %% Checking reconstructed image. %% % Using rsos (root sum of squares) to make visualization possible. dsImage = rsos(dsBrainCoils); % RSOS 重建得到 实值欠采样空间域混叠 MR 图像 120×128 double % figure; % imagesc(dsImage); % colormap('gray'); % colorbar(); figure; colormap('gray'); subplot(2, 3, 1); dsImage_1 = rsos(dsBrainCoils(:,:,1)) ; imagesc(dsImage_1); colormap('gray'); colorbar(); title('coil-1 partial FOV MR image'); subplot(2, 3, 2); dsImage_2 = rsos(dsBrainCoils(:,:,2)) ; imagesc(dsImage_2); colormap('gray'); colorbar(); title('coil-2 partial FOV MR image'); subplot(2, 3, 3); dsImage_3 = rsos(dsBrainCoils(:,:,3)) ; imagesc(dsImage_3); colormap('gray'); colorbar(); title('coil-3 partial FOV MR image'); subplot(2, 3, 4); dsImage_4 = rsos(dsBrainCoils(:,:,4)) ; imagesc(dsImage_4); colormap('gray'); colorbar(); title('coil-4 partial FOV MR image'); subplot(2, 3, 5); dsImage_5 = rsos(dsBrainCoils(:,:,5)) ; imagesc(dsImage_5); colormap('gray'); colorbar(); title('coil-5 partial FOV MR image'); subplot(2, 3, 6); imagesc(dsImage); title('RSOS reconstruction partial FOV MR image'); colorbar(); %% SENSE Reconstruction %% % Not a perfect solution but works for this experiment. fieldOfView = floor(phaseLength/downSamplingRate); % FOV_y = 120 / 5 = 24 % FOV_x = 128 senseRecon = zeros(phaseLength, freqLength); % 120×128 complex double - MR 重建结果 vectorI = zeros(coilNum, 1); % 5×1 complex double - 混叠像素向量(5个线圈在同一像素位置的复值) cHat = zeros(coilNum, downSamplingRate); % 5×5 double - 敏感度图(线圈数×重叠像素数) for x = 1:freqLength % kx = FOV_x = 128 for y = 1:fieldOfView % FOV_y = 24 % 欠采样 MR 图像在图像域 pixel(x, y) 处的 Nc=5 个混叠像素值存入 5×1 complex double 向量 I vectorI = reshape(dsBrainCoils(y, x, :), coilNum, 1); % All channels of single pixel in image. % 每个混叠像素 pixel(x, y) 都由 R = 5 个 desired full MR image 的像素重叠而成 for k = 1:downSamplingRate % R = 5 % Coil sensitivities of all channels of a single pixel per segment. cHat(:, k) = reshape(coilMap(y + (k-1) .* fieldOfView, x, :), coilNum, 1); % 5×1 double % y + (k-1).*fieldOfView 是敏感度图 FOV_y 方向上的偏移量 % cHat(:, k) 表示所有 5 个线圈在第 k 个 desired full MR image 像素点处的敏感度值 end % Eq.4 cHatPinv = pinv(cHat); % Moore-Penrose Pseudoinverse. vectorRho = cHatPinv * vectorI; % 5×1 complex double - 5 个 desired full MR image 像素点 for k = 1:downSamplingRate % 按 FOV_y 方向上的偏移量等距保存 desired full MR image 像素点 senseRecon(y + (k-1) .* fieldOfView, x) = vectorRho(k); end end end %% Visualizing results %% senseImage = rsos(senseRecon); senseImage = senseImage .* downSamplingRate; % Necessary to correct for the missing values. original = rsos(brainCoils); delta = original-senseImage; figure; colormap('gray'); subplot(2, 2, 1); imagesc(original); title('Original Image'); colorbar(); subplot(2, 2, 2); imagesc(dsImage); title('Downsampled Image'); colorbar(); subplot(2, 2, 3); imagesc(senseImage); title('SENSE Image'); colorbar(); subplot(2, 2, 4); imagesc(delta); title('Difference Image'); colorbar();参考链接:https://github.com/veritas9872/Medical-Imaging-Tutorial |
【本文地址】
今日新闻 |
推荐新闻 |