油菜根肿病抗性遗传改良与应用 |
您所在的位置:网站首页 › 杨梅新品种选育标准是什么 › 油菜根肿病抗性遗传改良与应用 |
油菜根肿病抗性遗传改良与应用
|
油菜根肿病抗性遗传改良与应用
王燕燕1,2,杨植全3,杨庆勇3,张椿雨1,2 1.华中农业大学国家油菜工程技术研究中心,武汉430070; 2.华中农业大学植物科学与技术学院,武汉430070;3.华中农业大学信息学院,武汉430070 摘要 油菜是我国重要的油料作物,但近年受根肿病危害严重,发病面积66.7万hm2以上,已成为影响油菜安全生产的头号杀手。油菜根肿病防治的方法较多,但最经济最有效的途径是抗性遗传改良。为了加快我国油菜根肿病抗性遗传改良的步伐,本文重点对抗性资源的发掘、抗性基因定位、抗病育种主要进展进行了总结,并适时提出一些尚待解决的科学或产业问题。由于长期种植单一抗性的油菜品种容易导致抗性丧失,本文还提出了持久抗性品种选育的可能途径,特别是对如何科学应用抗病品种给出了建议。 关键词 油菜; 根肿病; 抗性; 遗传改良; 抗病育种; 持久抗性; 基因定位; 种质资源; 聚合选育 油菜是我国重要的油料作物,年种植面积约666.7万hm2,产菜籽油500万t,约占自产食用植物油总量的50%,另提供约800万t的饲料饼粕[1]。近年来我国油菜生产饱受根肿病困扰。根肿病是由鞭毛菌亚门根肿菌属芸薹根肿菌(Plasmodiophora brassicae,为原生生物)侵染引起并专性为害十字花科植物(如油菜)的一种世界性土传病害。据不完全统计,我国油菜根肿病发病面积约66.7万hm2以上,主要集中在四川、湖北、湖南、云南和安徽等省的不同区域。发病区域油菜严重减产或绝收,并且随着我国农业机械化程度的不断提高,该土传病害正向全国油菜产区呈暴发式蔓延态势,已成为影响我国油菜产业安全生产的头号杀手。生产上防控根肿病的方式主要有农业防治(轮作、净土育苗移栽、迟播和施用石灰氮等)[2-5]、化学防控、生物防控和应用抗性品种等。这些措施中,比较有效的是净土育苗移栽、迟播和施用石灰氮。但净土育苗移栽通常费时费工、迟播可能会对适时播种产生不良影响,石灰氮由于价格较高、增加投入难以推广应用;此外,四川病区采取低温(气温连续低于15 ℃)条件下对叶面喷施一定浓度的生根剂,以促进新根生长从而挽救发病油菜植株,有一定的效果;但其中最有效、最经济的策略是抗性品种的选育与应用[6]。 1 根肿病抗性资源的发掘2019年以前关于油菜根肿病抗性资源发掘方面的研究进展总结于文献[6] 。总体而言,十字花科植物虽然种类繁多,但从中筛选到的抗性资源却寥寥无几。最新发表的质量性状抗源主要来自白菜类材料(AA,2n=20)[7-10]、芥菜型油菜(AABB,2n=36)[11]和萝卜(RR,2n=18)[12] 等。白菜型和芥菜型油菜抗性资源可通过与甘蓝型油菜(AACC,2n=38)直接进行有性杂交的方式而将抗性位点导入甘蓝型油菜。而萝卜与甘蓝型油菜杂交很困难,需要借助胚挽救及后续利用甘蓝型油菜为回交亲本进行不断回交方可成功。通过远缘杂交转育抗性基因的同时,往往有负面影响的连锁累赘性状也可同时被导入受体亲本[13]。目前表现为数量性状的抗源主要来自甘蓝型油菜[14-15]。质量性状抗性一般都有特异性,即对特定根肿菌生理小种类型有抗性,因此,长期种植含单一质量性状位点的抗性品种容易丧失抗性;而对于甘蓝型油菜含有的数量抗性材料,在根肿菌浓度较低的田块都表现出非常好的耐病性,而一旦在高浓度根肿病污染的田块其抗性一般都会表现很差。因此,本文重点关注基于质量性状抗性的甘蓝型油菜遗传改良研究。 然而,令人不解的是作为古老的病原物(根肿菌的病原最早在1878年由俄国科学家Worolin发现并命名为芸薹根肿菌),根肿菌在自然界中长期存在,但在种类和数量繁多的十字花科植物中仅找到数量极其有限的抗源材料,这是一个值得深入研究的重要科学问题。但只有待众多抗病基因被定位和克隆后,才可能从根本上揭示抗性资源材料的演化历程。 2 抗性基因的定位截至目前,国内外在含A基因组的不同抗性材料定位的质量抗性位点有26个(表1)。除了A基因组抗性材料外,近期在萝卜和芥菜型油菜中也分别定位了5个和1个抗性位点,其中芥菜型的抗性位点Rcr6与A基因组中定位的Crr1基因极可能为同源基因[11-12]。显然,在这些A基因组材料中定位的抗性位点也一定存在较多重叠。目前已经完成基因组测序的A基因组植物对根肿病均表现为感病,因此,缺乏抗病参考基因组,导致无法对这些不同抗性位点进行有效地归类、整合和抗性候选基因克隆,给合理利用上述抗源材料开展油菜根肿病抗性遗传改良研究带来较大困难。目前这些位点中只有CRa与Crr1a两个基因被成功克隆,均编码TIR-NB-LRR结构蛋白,为典型的R基因类型,但由上述抗性蛋白介导的抗病机制尚不清楚,其中Crr1a主要在根部、子叶、下胚轴等部位表达[29]。以含多个抗病位点的A基因组材料芜菁(如ECD04等)为研究对象,在完成基因组测序的基础上进行抗病位点整合和抗病候选基因克隆是未来研究的重点工作之一,也可为抗病机制的研究奠定重要基础。 表1 在A基因组不同抗源材料中定位或克隆的根肿病抗性位点及染色体分布Table 1 Clubroot resistance loci and chromosomal distribution located or cloned from different resistance sources in genome A 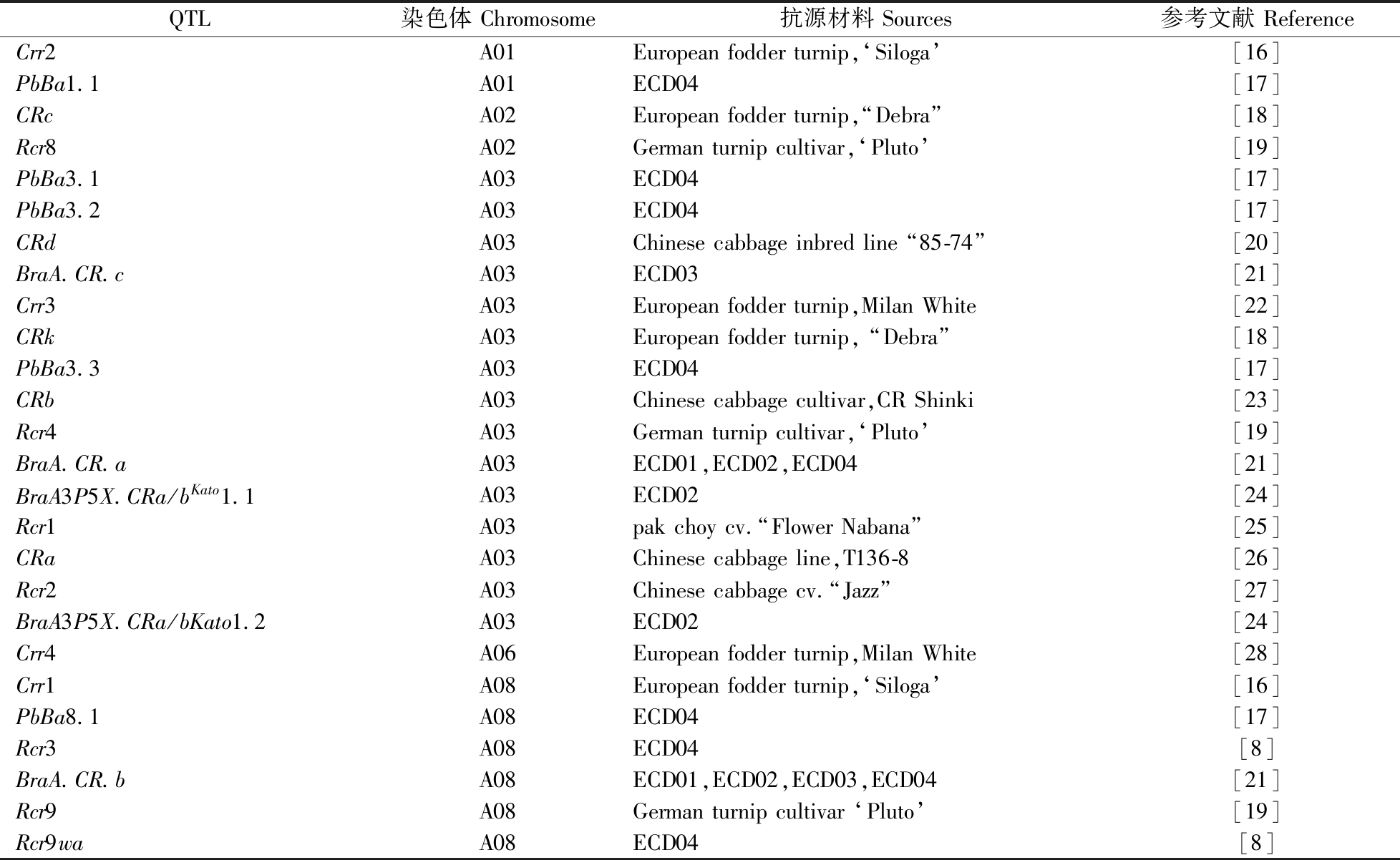
QTL染色体Chromosome抗源材料Sources参考文献ReferenceCrr2A01Europeanfodderturnip,‘Siloga’[16]PbBa1.1A01ECD04[17]CRcA02Europeanfodderturnip,“Debra”[18]Rcr8A02Germanturnipcultivar,‘Pluto’[19]PbBa3.1A03ECD04[17]PbBa3.2A03ECD04[17]CRdA03Chinesecabbageinbredline“85-74”[20]BraA.CR.cA03ECD03[21]Crr3A03Europeanfodderturnip,MilanWhite[22]CRkA03Europeanfodderturnip,“Debra”[18]PbBa3.3A03ECD04[17]CRbA03Chinesecabbagecultivar,CRShinki[23]Rcr4A03Germanturnipcultivar,‘Pluto’[19]BraA.CR.aA03ECD01,ECD02,ECD04[21]BraA3P5X.CRa/bKato1.1A03ECD02[24]Rcr1A03pakchoycv.“FlowerNabana”[25]CRaA03Chinesecabbageline,T136-8[26]Rcr2A03Chinesecabbagecv.“Jazz”[27]BraA3P5X.CRa/bKato1.2A03ECD02[24]Crr4A06Europeanfodderturnip,MilanWhite[28]Crr1A08Europeanfodderturnip,‘Siloga’[16]PbBa8.1A08ECD04[17]Rcr3A08ECD04[8]BraA.CR.bA08ECD01,ECD02,ECD03,ECD04[21]Rcr9A08Germanturnipcultivar‘Pluto’[19]Rcr9waA08ECD04[8] 3 甘蓝型油菜根肿病抗性育种种间杂交是转移有利性状的有效途径。因此,通过定向转育方法将根肿病抗性基因导入油菜是选育抗病品种的最有效途径。第1代抗根肿病油菜品种‘Mendel’和‘Tosca’是利用ECD04(芜菁,A基因组材料)和ECD15(甘蓝,CC,2n=18)种间杂交和染色体加倍,在创制出新型抗根肿病甘蓝型油菜资源的基础上选育出来[30]。随后,由于抗性资源被不断发掘与抗病基因定位工作的不断深入,油菜抗根肿病育种也取得了较好进展。加拿大油菜生产也同样受根肿病严重危害,截至2020年,Bayer、Pioneer和Cargill等9个大型种业公司共选育了53个抗根肿病新品种(https://www.canolacouncil.org/canola-encyclopedia/diseases/clubroot/),但这些品种是利用什么抗病位点尚无信息可查。我国油菜根肿病发生较加拿大晚,因此,根肿病抗病育种工作开展得也相对晚。华中农业大学通过将甘蓝型油菜优良亲本分别与抗病芜菁ECD04和抗病大白菜开展种间杂交,选育了我国首批抗根肿病常规新品种华双5R和杂交新品种华油杂62R并于2018年完成登记 [13,31-32]。这2个抗病品种分别含PbBa8.1和CRb两个抗病位点,对我国根肿菌4号优势生理小种均有免疫抗性,其中华油杂62R累计推广约3万~5万 hm2 [32]。同时,上述抗源材料分发给全国各主要油菜育种单位后,也带动了我国油菜抗根肿病新品种(如圣光165R、华油杂19R和华油杂160R等)的选育。随后,Shah等[33]系统研究了由这2个抗病位点衍生出的含单个抗病位点纯合、杂合,以及含2个抗病位点纯、杂合的6种基因型材料对云南腾冲、保山、临沧、楚雄,四川,湖北枝江、巴东,安徽黄山,辽宁新民等地根肿菌的抗病反应,结果表明:抗病纯合基因型的抗性强于杂合基因型,2个基因的抗性优于单个抗性基因,说明不同抗病基因的抗病反应可能存在互作关系,或者2个抗病位点存在剂量效应。前人研究也发现了类似现象,如Crr1对根肿菌的抗性表现为不完全显性,只有纯合时才对致病性较弱的菌株有抗性,Crr2单独存在时不对任何生理小种起抗性作用,但是当Crr1和Crr2同时存在时则对致病性较强的菌株表现出抗性[16,28,34]。从根肿菌的类型看, 四川、安徽、湖北等省的油菜主产区的变异较小,云南根肿菌多样性高,辽宁(新民)的根肿菌变异最大且与其他地区来源的菌株差别较大。以上研究结果的取得为我国油菜根肿病抗病育种指明了方向。 鉴于长期种植含单个抗病位点的油菜品种很容易造成抗性的丧失,未来油菜根肿病抗病育种的方向主要还是以多个抗性位点的分子聚合育种为主,这是因为抗病材料芜菁ECD04对来自我国不同地区的根肿菌具免疫抗性,其基因组中至少含3个或以上的抗性位点[17,21],而其中的抗病位点PbBa8.1只对部分地区的根肿菌存在免疫抗性。相信随着抗病资源及其抗病位点发掘的越来越多,运用与抗性位点紧密连锁的分子标记进行聚合选育具有持久抗性的油菜品种是未来工作的重点。 4 根肿病抗性油菜品种的应用为了更好地利用抗病品种并最大限度延长其使用年限,建议在推广和应用根肿病抗性油菜品种的过程中要重点关注以下问题:(1)根肿病的重发区以种植适宜的抗病品种为主。(2)在由政府采购并统一供种的地区,在对含不同抗病位点的品种有充分选择余地的前提下,农业管理部门可按年度交替使用含不同抗病位点的抗性品种,以适当延缓油菜品种抗性丧失的速度。(3)将抗病品种与10%左右的感病品种混合种植,可在一定程度上提高油菜群体抗病性的同时降低抗病品种抗性丧失的速度。最近研究表明,一定比例的抗、感病油菜品种混播,有利于提高群体的抗性水平[35];现有观点认为,抗病品种混有一定比例的感病品种,让感病品种正常发病,这可能既有利于保持根肿菌的种群稳定,还可减少根肿菌生理小种的变异速度,也不会因为种植密度发生较小的改变而影响最终产量。当然,这一观点尚缺乏有力证据支撑。为了获得有力证据,建议通过在根肿病发生区域对不同抗、感比例混合油菜品种进行长期定位种植,通过观察不同混合比例的抗、感油菜组合中抗病油菜抗性丧失的快慢及根肿菌变异情况来确定。(4)除在根肿病重发区种植抗病品种外,还建议在重发区周围的轻发区和未发病区域大面积种植抗病品种,以控制根肿病的进一步发展。 5 存在的问题及展望目前我国油菜根肿病防控过程中存在的主要问题是:由于根肿病随流水和机械作业传播速度快,加之根肿菌休眠孢子可以在土壤中存活20 a左右,且市场上可供选择的适合不同生态区种植的抗病品种还比较少,因此,预测我国油菜根肿病发病面积会越来越大并必将成为影响我国油菜安全生产的严重威胁。油菜根肿病抗性的遗传改良也必将成为我国油菜育种的核心和必需目标。 参考文献 References [1] 刘成,冯中朝,肖唐华,等.我国油菜产业发展现状、潜力及对策[J].中国油料作物学报,2019,41(4):485-489.LIU C,FENG Z C,XIAO T H,et al.Development,potential and adaptation of Chinese rapeseed industry[J].Chinese journal of oil crop sciences,2019,41(4):485-489(in Chinese with English abstract). [2] PENG G,STRELKOV S E,GROSSEN B D,et al.A >2-year crop rotation reduces resting spores of Plasmodiophora brassicae in soil and the impact of clubroot on canola[J].Eur J Agron,2015,70:78-84. [3] 唐利平,谢培庚,黄益国,等.油菜根肿病的发生及综合防控措施[J].湖南农业科学, 2019(7):65-67.TANG L P,XIE P G,HUANG Y G,et al.Occurrence of Brassica napus root swollen disease and comprehensive prevention and control measures[J].Hunan agricultural sciences,2019(7):65-67(in Chinese with English abstract). [4] 费维新,王淑芬,李强生,等.冬油菜适当迟播有效减轻油菜根肿病[J].中国油料作物学报,2016,38(4):502-507.FEI W X,WANG S F,LI Q S,et al.Reducing clubroot disease by late sowing of winter rapeseed[J].Chinese journal of oil crop sciences,2016,38(4):502-507(in Chinese with English abstract). [5] NIWA R,KUMEI T,NOMURA Y,et al.Increase in soil pH due to Ca-rich organic matter application causes suppression of the clubroot disease of crucifers[J].Soil Biol Bio-chem,2007,39:778-785. [6] 江莹芬,战宗祥,朴钟云,等.油菜抗根肿病资源创新与利用的研究进展与展望[J].作物学报,2018,44(11):1592-1599.JIANG Y F,ZHAN Z X,PIAO Z Y,et al.Progresses and prospects of germplasms innovation for clubroot resistance and genetic improvement in Brassica napus[J].Acta agronomic asinica,2018,44(11):1592-1599(in Chinese with English abstract). [7] HUANG Z,PENG G,GOSSEN B D,et al.Fine mapping of a clubroot resistance gene from turnip using SNP markers identified from bulked segregant RNA-Seq[J/OL].Molecular breeding,2019,39(9):131[2021-02-16].https://doi.org/10.1007/s11032-019-1038-8. [8] KARIM M M,DAKOURI A,ZHANG Y,et al.Two clubroot-resistance genes,Rcr3 and Rcr9wa,mapped in Brassica rapa using bulk segregant RNA sequencing[J/OL].International journal of molecular sciences,2020,21(14):5033[2021-02-16].https://doi.org/10.3390/ijms21145033. [9] LAILA R,PARK J I,ROBIN A H K,et al.Mapping of a novel clubroot resistance QTL using ddRAD-seq in Chinese cabbage ( Brassica rapa L.)[J/OL].BMC plant biology,2019,19(1):13[2021-02-16].https://doi.org/10.1186/s12870-018-1615-8. [10] CHOI S R,OH S H,CHHAPEKAR S S,et al.Quantitative trait locus mapping of clubroot resistance and Plasmodiophora brassicae pathotype banglim-specific marker development in Brassica rapa[J/OL].International journal of molecular sciences,2020,21(11):4157[2021-02-16].https://doi.org/10.3390/ijms21114157. [11] CHANG A,LAMARA M,WEI Y,et al.Clubroot resistance gene Rcr6 in Brassica nigra resides in a genomic region homologous to chromosome A08 in B. rapa[J/OL].BMC plant biology,2019,19(1):224[2021-02-16].https://doi.org/10.1186/s12870-019-1844-5. [12] GAN C X,DENG X H,CUI L,et al.Construction of a high-density genetic linkage map and identification of quantitative trait loci associated with clubroot resistance in radish (Raphanus sativus L.)[J].Molecular breeding,2019,39:116-127. [13] ZHAN Z,SHAH N,ZHANG C.Association of clubroot resistance locus PbBa8.1 with a linkage drag of high erucic acid content in the seed of the European turnip[J/OL].Frontiers in plant science,2020,11:810[2021-02-16].https://doi.org/10.3389/fpls.2020.00810. [14] HASAN M J,RAHMAN M H.Genetics and molecular mapping of resistance to Plasmodiophora brassicae pathotypes 2,3,5,6 and 8 in rutabaga (Brassica napus var. napobrassica)[J].Genome,2016,59(10):805-815. [15] LI L,LUO Y,CHEN B,et al.Genome-wide association study reveals new loci for resistance to clubroot disease in Brassica napus[J/OL].Frontiers in plant science,2016,7(21625):[2021-02-16].https://doi.org/10.3389/fpls.2016.01483. [16] SUWABE K,TSUKAZAKI H,IKETANI H,et al.Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L.[J].Theoretical & applied genetics,2003,107(6):997-1002. [17] CHEN J,JING J,ZHAN Z,et al.Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa[J/OL].PLoS One, 2013,8(12):e85307[2021-02-16].https://doi.org/10.1371/journal.pone.0085307. [18] SAKAMOTO K,SAITO A,HAYASHIDA N,et al.Mapping of isolate-specific QTL for clubroot resistance in Chinese cabbage (Brassica rapa L.ssp. pekinensis)[J].Theoretical and applied genetics,2008,117(5):759-767. [19] YU F,ZHANG X,PENG G,et al.Genotyping-by-sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa[J/OL].Rep,2017,7(1):4516[2021-02-16].https://doi.org/10.1038/s41598-017-04903-2. [20] PENG W X,FU P Y,LI X N,et al.Identification and mapping of the clubroot resistance gene crd in chinese cabbage (Brassica rapa ssp.pekinensis)[J/OL].Frontiers in plant science, 2018,9:653[2021-02-16].https://doi.org/10.3389/fpls.2018.00653. [21] HIRANI A H,GAO F,LIU J,et al.Combinations of independent dominant loci conferring clubroot resistance in all four turnip accessions (Brassica rapa) from the European clubroot differential set[J/OL].Frontiers in plant science,2018,9:1628[2021-02-16].https://doi.org/10.3389/fpls.2018.01628. [22] HIRAI M,HARADA T,KUBO N,et al.A novel locus for clubroot resistance in Brassica rapa and its linkage markers[J].Theoretical & applied genetics, 2004,108(4):639-643. [23] PIAO Z Y,DENG Y Q,CHOI S R,et al.SCAR and CAPS mapping of CRb,a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage ( Brassica rapa ssp. pekinensis)[J].Theoretical & applied genetics,2004,108(8):1458-1465. [24] FREDUA-AGYEMAN R,JIANG J,HWANG S F,et al.QTL mapping and inheritance of clubroot resistance genes derived from Brassica rapa subsp. rapifera (ECD 02) reveals resistance loci and distorted segregation ratios in two F2 populations of different crosses[J/OL].Frontiers in plant science,2020,11:899[2021-02-16].https://doi.org/10.3389/fpls.2020.00899. [25] CHU M,SONG T,FALK K C,et al.Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae[J/OL].BMC genomics, 2014,15(1):1166[2021-02-16].https://doi.org/10.1186/1471-2164-15-1166. [26] MATSUMOTO E,YASUI C,OHI M,et al.Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp.pekinensis)[J].Euphytica,1998,104(2):79-86. [27] HUANG Z,PENG G,GOSSEN B D,et al.Fine mapping of a clubroot resistance gene from turnip using SNP markers identified from bulked segregant RNA-Seq[J/OL].Molecular breeding,2017,8:1448[2021-02-16].https://doi.org/10.3389/fpls.2017.01448. [28] SUWABE K.Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana:the genetic origin of clubroot resistance[J].Genetics,2006,173(1):309-319. [29] KATSUNORI H,KEITA S,NORIO T R,et al.Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L.[J/OL].PLoS One,2013,8(1):e54745[2021-02-16].https://doi.org/10.1371/journal.pone.0054745. [30] DIEDERICHSEN E,SACRISTAN M D.Disease response of resynthesized Brassica napus L. lines carrying different combinations of resistance to Plasmodiophora brassicae Wor[J].Plant breeding,2006,115(1):5-10. [31] 战宗祥,江莹芬,朱紫媛,等.与位点PbBa8.1紧密连锁分子标记的开发及甘蓝型油菜根肿病抗性育种[J].中国油料作物学报,2015,37(6) :766-771.ZHAN Z X,JIANG Y F,ZHU Z Y,et al.Development of close linked marker to PbBa8.1 conferring canola resistance to Plasmodiophora brassicae[J].Chinese journal of oil crop sciences,2015,37(6) :766 -771(in Chinese with English abstract). [32] 李倩,NADIL S,周元委,等.抗根肿病甘蓝型油菜新品种华油杂 62R 的选育[J].作物学报,2021,47(2):210-223.LI Q,NADIL SHAH,ZHOU Y W,et al.Breeding of a novel clubroot disease-resistant Brassica napus variety Huayouza 62R[J].Acta agronomic asinica,2021,47(2):210-223(in Chinese with English abstract). [33] SHAH N,SUN J,YU S,et al.Genetic variation analysis of field isolates of clubroot and their responses to Brassica napus lines containing resistant genes CRb and PbBa8.1 and their combination in homozygous and heterozygous state[J/OL].Molecular breeding 2019,39(10/11):153[2021-02-16].https://doi.org/10.1007/s11032-019-1075-3. [34] DEAN A.On a chromosome far,far away:LCRs and gene expression[J].Trends in genetics,2006,22(1):38-45. [35] 郭清云,汪波,蒯婕,等.油菜感抗品种混播对根肿病防控效果的影响[J].作物学报,2020,46(5):725-733.GUO Q Y,WANG B,KUAI J,et al.Controlling efficiency against clubroot disease of rapeseed by mixed-cropping of susceptible and resistant cultivars[J].Acta agronomic asinica,2020,46(5):725-733(in Chinese with English abstract). Genetic improvement and application of clubroot resistance in Brassica napus varietiesWANG Yanyan1,2,YANG Zhiquan3,YANG Qingyong3,ZHANG Chunyu1,2 1.National Research Center of Rapeseed Engineering and Technology,Huazhong Agricultural University,Wuhan 430070,China; 2.College of Plant Science and Technology,Huazhong Agricultural University,Wuhan 430070,China;3.College of Information,Huazhong Agricultural University,Wuhan 430070,China Abstract Rapeseed is the largest oil crop in China. However,it has been seriously affected by clubroot disease caused by Plasmodiophora brassicae in recent years,with the infected area of more than 667 000 hm2. The clubroot disease has become the number one killer affecting the safety of rapeseed production. There are many methods for prevention and treatment of clubroot disease in rapeseed,but the most economical and effective way is the genetic improvement of disease resistance. In order to accelerate the genetic improvement of clubroot disease resistance in rapeseed in China,this article summarized the progress in discovering resources of disease resistance,mapping genes of disease resistance,and breeding of disease resistance. Some scientific or industrial problems to be resolved are put forward in a timely manner. Since long-term planting of rapeseed varieties with single resistance can easily lead to the loss of resistance,this article proposes possible ways to breed varieties with long-lasting resistance. The specific suggestions on how to develop rapeseed varieties with durable resistance and how to utilize them properly in the field are provided. Keywords Brassica napus; clubroot; resistance; genetic improvement; breeding of disease resistance; durable resistance; gene mapping; germplasm resources; aggregation breeding 中图分类号 S 565.403.4 文献标识码 A 文章编号 1000-2421(2021)02-0001-05 王燕燕,杨植全,杨庆勇,等.油菜根肿病抗性遗传改良与应用[J].华中农业大学学报,2021,40(2):1-5. DOI:10.13300/j.cnki.hnlkxb.2021.02.001 收稿日期: 2021-02-16 基金项目:国家自然科学基金项目(31871659);国家油菜产业技术体系(CARS-12) 王燕燕,E-mail:[email protected] 通信作者:张椿雨,E-mail:[email protected] (责任编辑:张志钰) |
【本文地址】