|
文章目录
一、什么是参考基因组和基因组注释?二、参考基因组版本命名1、常用人参考基因组对应表2、常用小鼠参考基因组对应表
三、下载1、NCBI2、Ensembl3、GENCODE4、UCSC5、iGenomes
四、其他参考基因组信息
一、什么是参考基因组和基因组注释?
先来理一理参考基因组,基因组注释文件间的关系。
自从 1990 启动的家喻户晓的人类基因组计划开始,全世界的科学家竭尽全力破译了第一个完整的人类基因组,从那时开始人类拿到了一本只有 ATCG 四个碱基书写的天书。后续人们逐步完善了基因组序列信息,并写在 Fasta 格式的文本文件“天书”中,这本天书就叫做参考基因组。

但是,直接拿天书来看是一脸懵逼的,于是大家开始利用实验技术手段开始着手解密这本天书,随后大量的基因以及非编码序列被人们详细的标记在参考基因组对应的位置。同时对该位置加入大量的注释细节,最终将这些信息写在 BED,GTF,GFF 格式的基因组注释文件 。所以也可以把基因组注释文件理解为字典,看不懂天书,翻翻字典就懂了。
 随着时间的推移,在更先进技术的加持下,在已经构建好的基因组和注释信息上不断增加,删减,修改,就有了不同的版本。而每一个版本的参考基因组都会对应有一个基因组注释文件(天书和字典一一对应),接下来我们看看参考基因组版本是怎么指定的。 随着时间的推移,在更先进技术的加持下,在已经构建好的基因组和注释信息上不断增加,删减,修改,就有了不同的版本。而每一个版本的参考基因组都会对应有一个基因组注释文件(天书和字典一一对应),接下来我们看看参考基因组版本是怎么指定的。
二、参考基因组版本命名
在讲参考基因组之前,需要提到一个组织参考基因组联盟(Genome Reference Consortium),它是由 NCBI,EBI,桑格研究所等机构组成。GRC 利用最佳的技术装配,纠正,增加基因组序列,以此作为在生信分析领域作为参考的基因组。目前,该机构构建了人,小鼠,大鼠,斑马鱼,鸡的参考基因组。
人基因组官名叫 GRCh38 (Genome Reference Consortium Human Build 38),GRCh38 在UCSC基因组浏览器中还有个小名 hg38,这个小名对于大多数人来说是更亲切熟悉的。GRCh38 在 GenBank 中叫 GCA_000001405.15,在 RefSeq 中叫 GCF_000001405.26,虽然 GRC 组织建议在所有出版物和工具中使用该编号,但事实是前两种 GRCh38 和 hg38 对生信分析更常见。
在不更改染色体坐标的情况下,向参考基因组添加或替换新序列,这种打补丁的方式,会在基因组版本后加 .p (patch)来命名。
这就像在王者荣耀,英雄联盟中,为了维持游戏热度,会大幅修改游戏架构,流程,世界观,图片,叫大版本更新,而定期对某些英雄的面板属性修正,作为补丁。
举个例子,GRCh38 的第九个补丁,正式版本叫做 Genome Reference Consortium Human Build 38 patch release 9,简称 GRCh38.p9。在 GenBank 编号为 GCA_000001405.24,RefSeq 编号为 GCF_000001405.35。在 Ensemble 编号为 GRCh38,NCBI 编号为 GRCh38。
1、常用人参考基因组对应表
发布时间201320092006GRC 官名GRCh38GRCh37GRCh36UCSChg38hg19hg18EnsembleGRCh38GRCh37GRCh36GENCODE38193cNCBIGRCh38GRCh37GRCh36GenBankGCA_000001405RefSeqGCF_000001405
根据 GRC 官网信息,GRCh39 大版本将会无限停更,他们在考虑用新模型和序列来构建人类的参考基因组,细节不清楚,猜测有可能会有泛基因组内容。
2、常用小鼠参考基因组对应表
发布时间202020112007GRC 官名GRCm39GRCm38UCSCm39mm10mm9EnsembleGRCm39GRCm38GENCODEM27M25M1NCBIGRCm39GRCm38NCBIM37
三、下载
1、NCBI
这里提供两种下载方式,一种为网页界面下载,另一种为FTP下载。
可视化下载
进入网址
https://www.ncbi.nlm.nih.gov/genome/browse#!/overview/
搜索物种
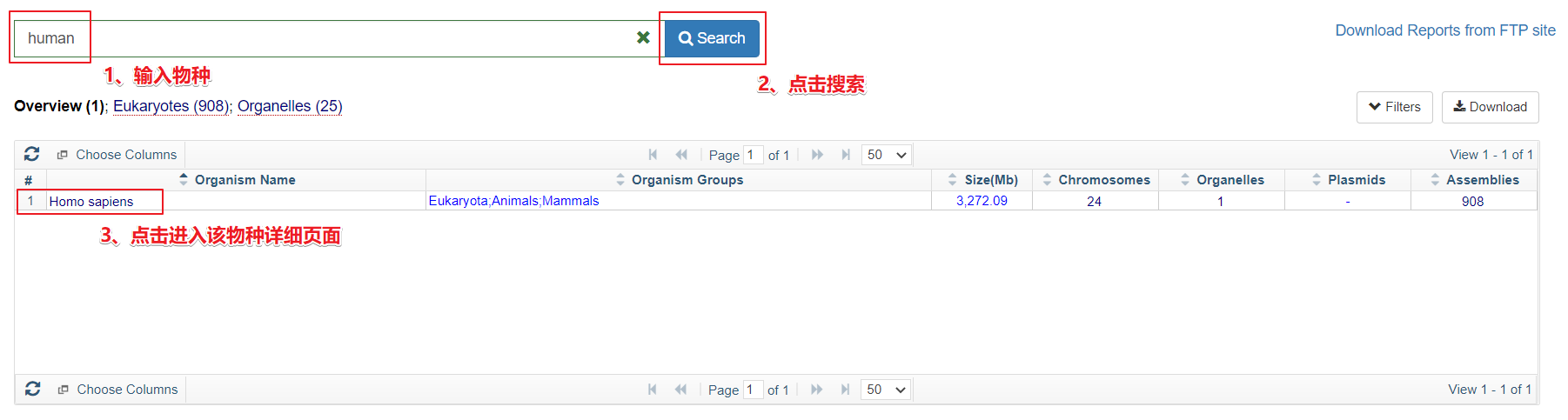
下载界面
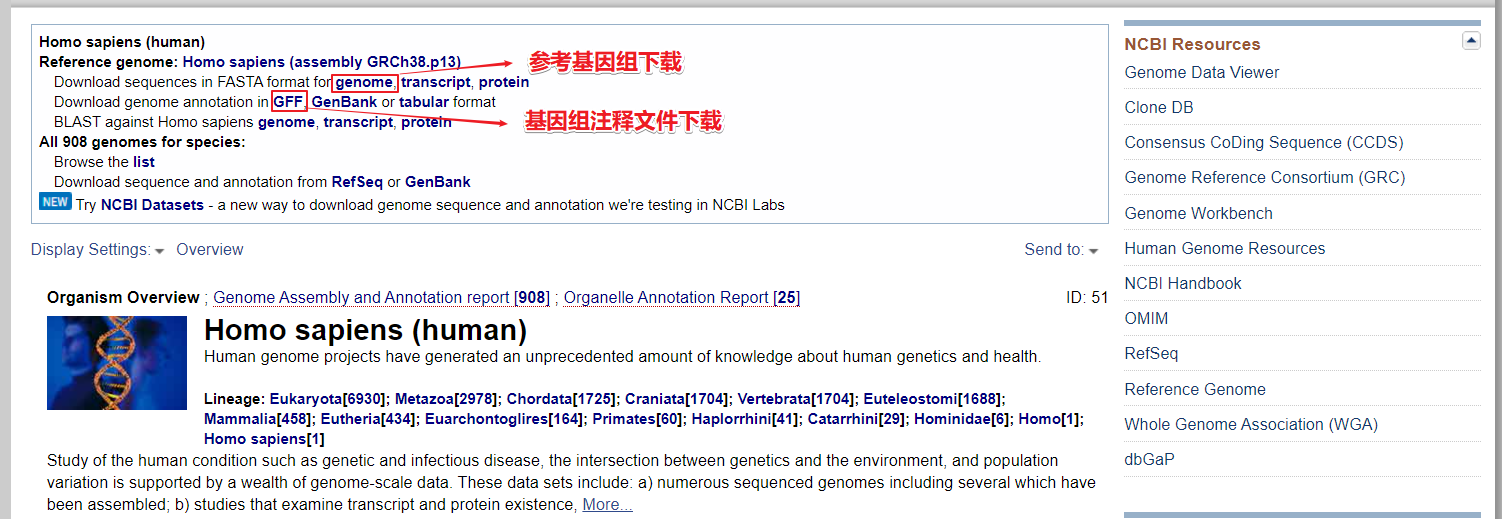
FTP下载
随便提一下,Chrome 浏览器在18版本后由于安全原因已经不支持 ftp 协议,改用 https 协议,可以看到链接已经与之前的不同。
这里以下载人的参考基因组 GRCh38 为例:
https://ftp.ncbi.nlm.nih.gov/genomes/refseq/vertebrate_mammalian/Homo_sapiens/reference/GCF_000001405.39_GRCh38.p13
人类基因组注释文件:
GTF 格式:https://ftp.ncbi.nlm.nih.gov/genomes/refseq/vertebrate_mammalian/Homo_sapiens/annotation_releases/109/GCF_000001405.38_GRCh38.p12/GCF_000001405.38_GRCh38.p12_genomic.gtf.gz
GFF 格式:
https://ftp.ncbi.nlm.nih.gov/genomes/refseq/vertebrate_mammalian/Homo_sapiens/annotation_releases/109/GCF_000001405.38_GRCh38.p12/GCF_000001405.38_GRCh38.p12_genomic.gff.gz
如果以这种方式下载,其实已经可以路径中大概看出相关物种的下载地址,可以自行查询及下载其他物种。
2、Ensembl
可视化下载
网址:http://asia.ensembl.org点击物种名,进入下载界面
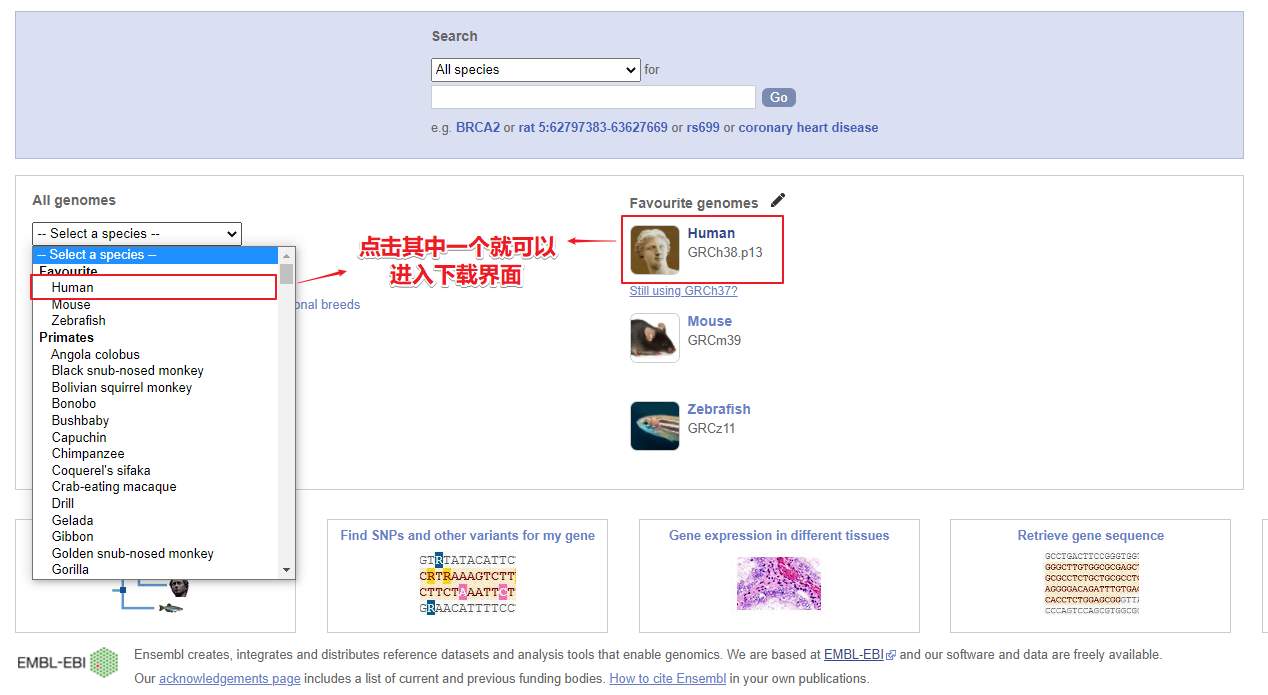
点击对应名称,下载参考基因组和基因组注释文件
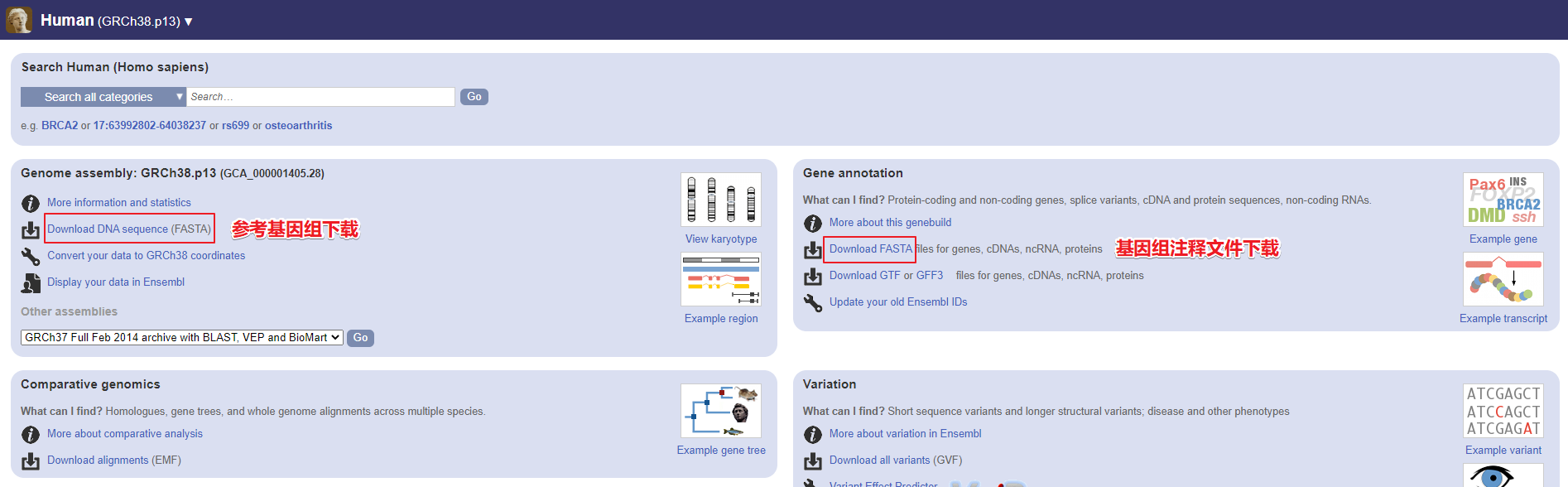
FTP下载
同样以下载人参考基因组 GRCh38 为例:
http://ftp.ensembl.org/pub/current_fasta/homo_sapiens/dna/Homo_sapiens.GRCh38.dna.toplevel.fa.gz
GTF 文件:http://ftp.ensembl.org/pub/current_gtf/homo_sapiens/Homo_sapiens.GRCh38.104.gtf.gz
GTT 文件:http://ftp.ensembl.org/pub/current_gff3/homo_sapiens/Homo_sapiens.GRCh38.104.gff3.gz
3、GENCODE
如果小伙伴研究的物种只涉及人类和小鼠,极力推荐 GENCOE,这里有着相较其他数据库,最新最全的基因组和其注释信息。
网址:https://www.gencodegenes.org/点击人类的最新版
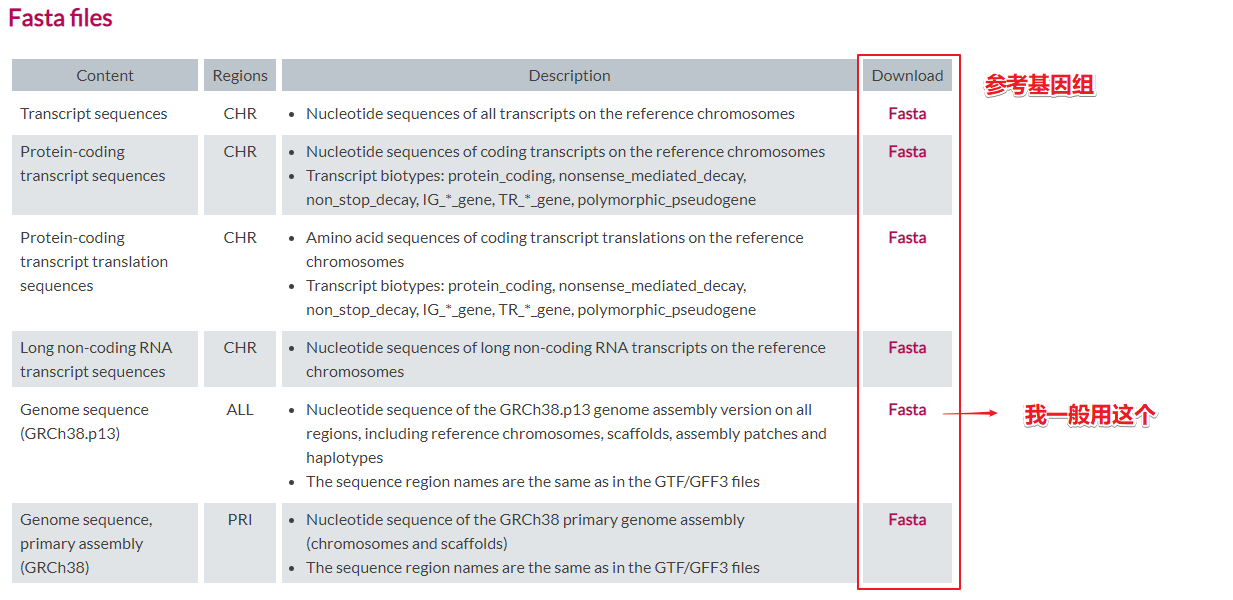
点击下载基因组注释文件

点击下载参考基因组文件
4、UCSC
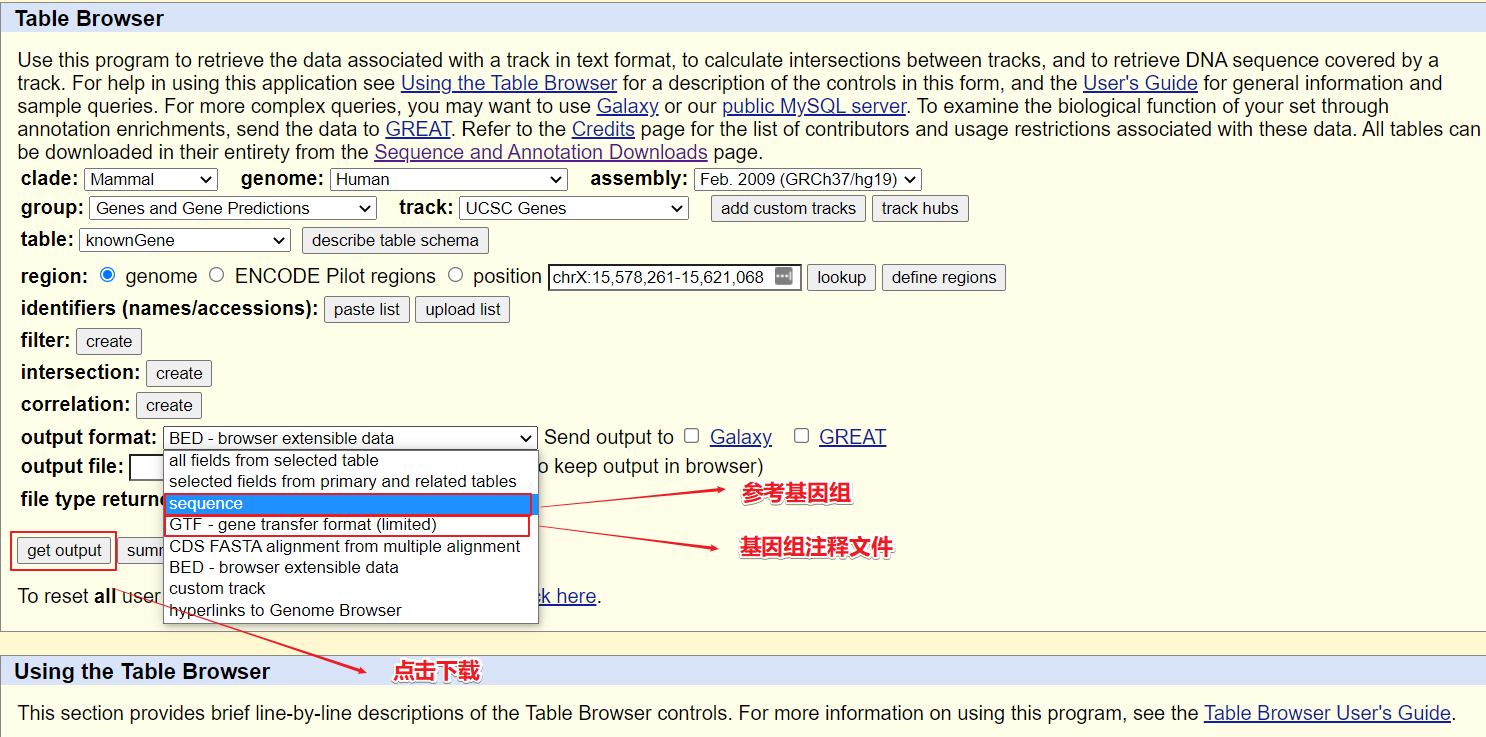
相对其他下载方式,UCSC 本职的工作是做基因组浏览器的,因此也可以从下图看到,在这里可以根据自己定义来下载相对于的基因组区域,比如 prime,exon,gene,transcript等等。
网址:http://genome.ucsc.edu/cgi-bin/hgTables下载:设置参数如下,然后点击下载参考基因组及注释文件
5、iGenomes
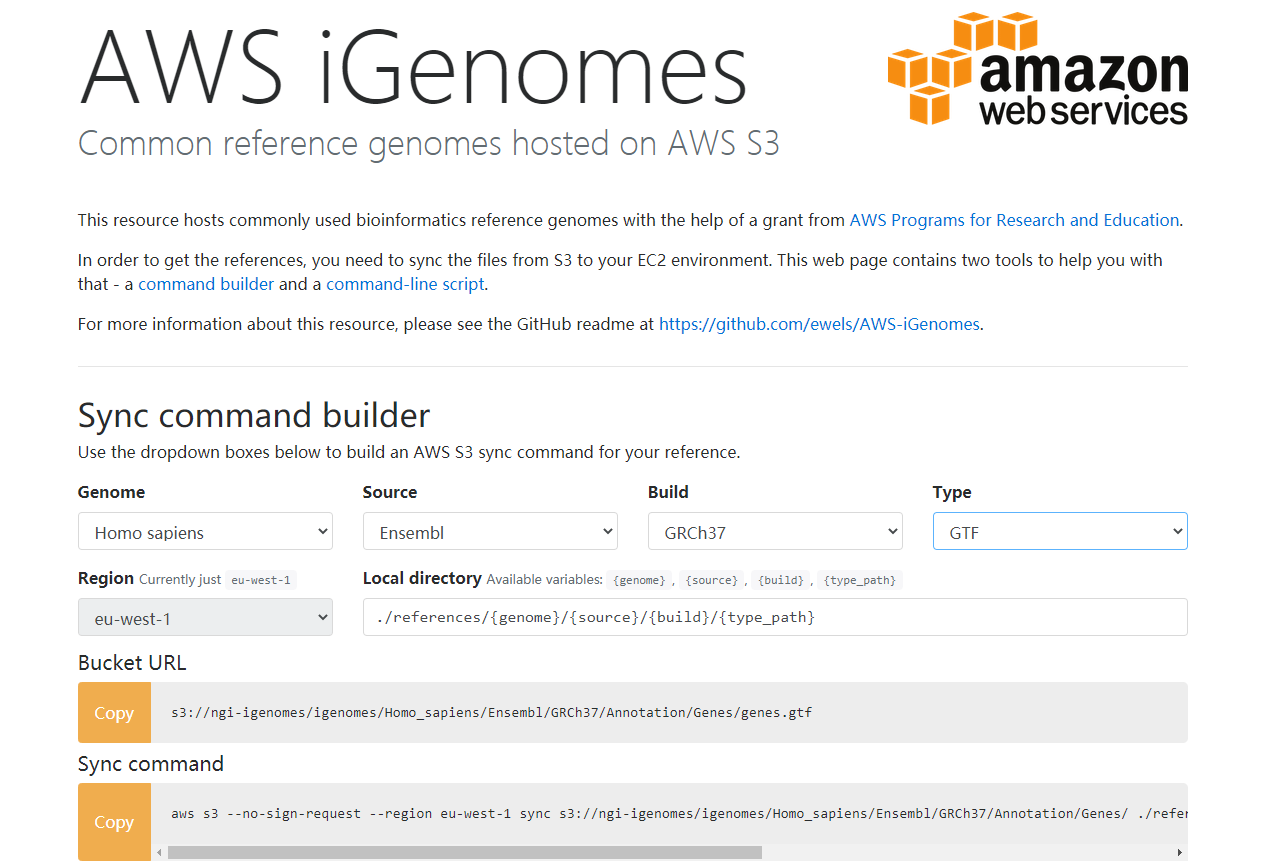
iGenomes是常见分析生物的参考序列和注释文件的集合。这些文件已从Ensembl,NCBI或UCSC下载。染色体名称已更改为简单且与下载源一致。每个iGenome都可以作为压缩文件使用,其中包含生物体的单个基因组构建的序列和注释文件。
网址:https://support.illumina.com/sequencing/sequencing_software/igenome.html
由亚马逊资助的生物信息参考基因组下载站点,有各种参考基因组,注释文件,软件索引等常用文件,并且有着极快的下载速度,但是缺点是只有常用的物种。
**站点:**https://ewels.github.io/AWS-iGenomes/
四、其他参考基因组信息
SPECIESUCSC VERSIONRELEASE DATERELEASE NAMESTATUSMAMMALSHumanhg38Dec. 2013Genome Reference Consortium GRCh38Availablehg19Feb. 2009Genome Reference Consortium GRCh37Availablehg18Mar. 2006NCBI Build 36.1Availablehg17May 2004NCBI Build 35Availablehg16Jul. 2003NCBI Build 34Availablehg15Apr. 2003NCBI Build 33Archivedhg13Nov. 2002NCBI Build 31Archivedhg12Jun. 2002NCBI Build 30Archivedhg11Apr. 2002NCBI Build 29Archived (data only)hg10Dec. 2001NCBI Build 28Archived (data only)hg8Aug. 2001UCSC-assembledArchived (data only)hg7Apr. 2001UCSC-assembledArchived (data only)hg6Dec. 2000UCSC-assembledArchived (data only)hg5Oct. 2000UCSC-assembledArchived (data only)hg4Sep. 2000UCSC-assembledArchived (data only)hg3Jul. 2000UCSC-assembledArchived (data only)hg2Jun. 2000UCSC-assembledArchived (data only)hg1May 2000UCSC-assembledArchived (data only)AlpacavicPac2Mar. 2013Broad Institute Vicugna_pacos-2.0.1AvailablevicPac1Jul. 2008Broad Institute VicPac1.0AvailableArmadillodasNov3Dec. 2011Broad Institute DasNov3AvailableBaboonpapAnu4Apr. 2017Human Genome Sequencing CenterAvailablepapAnu2Mar. 2012Baylor College of Medicine Panu_2.0AvailablepapHam1Nov. 2008Baylor College of Medicine HGSC Pham_1.0AvailableBisonbisBis1Oct. 2014Univ. of Maryland Bison_UMD1.0AvailableBonobopanPan3May 2020University of WashingtonAvailablepanPan2Dec. 2015Max-Planck Institute for Evolutionary Anthropology panpan1.1AvailablepanPan1May 2012Max-Planck Institute panpan1AvailableBrown kiwiaptMan1Jun. 2015Max-Planck Institute for Evolutionary Anthropology AptMant0AvailableBushbabyotoGar3Mar. 2011Broad Institute OtoGar3AvailableCatfelCat9Nov. 2017Genome Sequencing Center (GSC) at Washington University (WashU) School of Medicine Felis_catus_9.0AvailablefelCat8Nov. 2014ICGSC Felis_catus_8.0AvailablefelCat5Sep. 2011ICGSC Felis_catus-6.2AvailablefelCat4Dec. 2008NHGRI catChrV17eAvailablefelCat3Mar. 2006Broad Institute Release 3AvailableChimppanTro6Jan. 2018Clint_PTRv2AvailablepanTro5May 2016CGSC Build 3.0AvailablepanTro4Feb. 2011CGSC Build 2.1.4AvailablepanTro3Oct. 2010CGSC Build 2.1.3AvailablepanTro2Mar. 2006CGSC Build 2.1AvailablepanTro1Nov. 2003CGSC Build 1.1AvailableChinese hamstercriGri1Jul. 2013Beijing Genomics Institution-Shenzhen C_griseus_v1.0AvailableChinese hamster ovary cell linecriGriChoV2Jun. 2017Eagle Genomics Ltd CHOK1S_HZDv1AvailablecriGriChoV1Aug. 2011Beijing Genomics Institute CriGri_1.0AvailableChinese pangolinmanPen1Aug. 2014Washington University (WashU) M_pentadactyla-1.1.1AvailableCowbosTau9Apr. 2018USDA ARSAvailablebosTau8Jun. 2014University of Maryland v3.1.1AvailablebosTau7Oct. 2011Baylor College of Medicine HGSC Btau_4.6.1AvailablebosTau6Nov. 2009University of Maryland v3.1AvailablebosTau4Oct. 2007Baylor College of Medicine HGSC Btau_4.0AvailablebosTau3Aug. 2006Baylor College of Medicine HGSC Btau_3.1AvailablebosTau2Mar. 2005Baylor College of Medicine HGSC Btau_2.0AvailablebosTau1Sep. 2004Baylor College of Medicine HGSC Btau_1.0ArchivedCrab-eating macaquemacFas5Jun. 2013Washington University Macaca_fascicularis_5.0AvailableDogcanFam5May 2019University of MichiganAvailablecanFam4Mar. 2020Uppsala UniversityAvailablecanFam3Sep. 2011Broad Institute v3.1AvailablecanFam2May 2005Broad Institute v2.0AvailablecanFam1Jul. 2004Broad Institute v1.0AvailableDolphinturTru2Oct. 2011Baylor College of Medicine Ttru_1.4AvailableElephantloxAfr3Jul. 2009Broad Institute LoxAfr3AvailableFerretmusFur1Apr. 2011Ferret Genome Sequencing Consortium MusPutFur1.0AvailableGarter snakethaSir1Jun. 2015Washington University Thamnophis_sirtalis-6.0AvailableGibbonnomLeu3Oct. 2012Gibbon Genome Sequencing Consortium Nleu3.0AvailablenomLeu2Jun. 2011Gibbon Genome Sequencing Consortium Nleu1.1AvailablenomLeu1Jan. 2010Gibbon Genome Sequencing Consortium Nleu1.0AvailableGolden eagleaquChr2Oct. 2014University of Washington aquChr2-1.0.2AvailableGolden snub-nosed monkeyrhiRox1Oct. 2014Novogene Rrox_v1AvailableGorillagorGor6Aug. 2019University of WashingtonAvailablegorGor5Mar. 2016University of Washington GSMRT3AvailablegorGor4Dec. 2014Wellcome Trust Sanger Institute gorGor4AvailablegorGor3May 2011Wellcome Trust Sanger Institute gorGor3.1AvailableGreen MonkeychlSab2Mar. 2014Vervet Genomics Consortium 1.1AvailableGuinea pigcavPor3Feb. 2008Broad Institute cavPor3AvailableHawaiian monk sealneoSch1Jun. 2017Johns Hopkins University ASM220157v1AvailableHedgehogeriEur2May 2012Broad Institute EriEur2.0AvailableeriEur1Jun. 2006Broad Institute Draft_v1AvailableHorseequCab3Jan. 2018University of LouisvilleAvailableequCab2Sep. 2007Broad Institute EquCab2AvailableequCab1Jan. 2007Broad Institute EquCab1AvailableKangaroo ratdipOrd1Jul. 2008Baylor/Broad Institute DipOrd1.0AvailableMalayan flying lemurgalVar1Jul. 2014WashU G_variegatus-3.0.2AvailableManateetriMan1Oct. 2011Broad Institute TriManLat1.0AvailableMarmosetcalJac4May 2020Washington University Callithrix_jacchus_cj1700_1.1AvailableMarmosetcalJac3Mar. 2009WUSTL Callithrix_jacchus-v3.2AvailablecalJac1Jun. 2007WUSTL Callithrix_jacchus-v2.0.2AvailableMegabatpteVam1Jul. 2008Broad Institute Ptevap1.0AvailableMicrobatmyoLuc2Jul. 2010Broad Institute MyoLuc2.0AvailableMinke whalebalAcu1Oct. 2013KORDI BalAcu1.0AvailableMousemm39Jun. 2020Genome Reference Consortium Mouse Build 39Availablemm10Dec. 2011Genome Reference Consortium GRCm38Availablemm9Jul. 2007NCBI Build 37Availablemm8Feb. 2006NCBI Build 36Availablemm7Aug. 2005NCBI Build 35Availablemm6Mar. 2005NCBI Build 34Archivedmm5May 2004NCBI Build 33Archivedmm4Oct. 2003NCBI Build 32Archivedmm3Feb. 2003NCBI Build 30Archivedmm2Feb. 2002MGSCv3Archivedmm1Nov. 2001MGSCv2Archived (data only)Mouse lemurmicMur2May 2015Baylor/Broad Institute Mmur_2.0AvailablemicMur1Jul. 2007Broad Institute MicMur1.0AvailableNaked mole-rathetGla2Jan. 2012Broad Institute HetGla_female_1.0AvailablehetGla1Jul. 2011Beijing Genomics Institute HetGla_1.0AvailableOpossummonDom5Oct. 2006Broad Institute release MonDom5AvailablemonDom4Jan. 2006Broad Institute release MonDom4AvailablemonDom1Oct. 2004Broad Institute release MonDom1AvailableOrangutanponAbe2Jul. 2007WUSTL Pongo_albelii-2.0.2AvailableponAbe3Jan. 2018Susie_PABv2/ponAbe3AvailablePandaailMel1Dec. 2009BGI-Shenzhen AilMel 1.0AvailablePigsusScr11Feb. 2017Swine Genome Sequencing Consortium Sscrofa11.1AvailablesusScr3Aug. 2011Swine Genome Sequencing Consortium Sscrofa10.2AvailablesusScr2Nov. 2009Swine Genome Sequencing Consortium Sscrofa9.2AvailablePikaochPri3May 2012Broad Institute OchPri3.0AvailableochPri2Jul. 2008Broad Institute OchPri2AvailablePlatypusornAna2Feb. 2007WUSTL v5.0.1AvailableornAna1Mar. 2007WUSTL v5.0.1AvailableProboscis MonkeynasLar1Nov. 2014Proboscis Monkey Functional Genome Consortium Charlie1.0AvailableRabbitoryCun2Apr. 2009Broad Institute release OryCun2AvailableRatrn7Nov. 2020Wellcome Sanger Institute mRatBN7.2Availablern6Jul. 2014RGSC Rnor_6.0Availablern5Mar. 2012RGSC Rnor_5.0Availablern4Nov. 2004Baylor College of Medicine HGSC v3.4Availablern3Jun. 2003Baylor College of Medicine HGSC v3.1Availablern2Jan. 2003Baylor College of Medicine HGSC v2.1Archivedrn1Nov. 2002Baylor College of Medicine HGSC v1.0ArchivedRhesusrheMac10Feb. 2019The Genome Institute at Washington University School of Medicine Mmul_10AvailablerheMac8Nov. 2015Baylor College of Medicine HGSC Mmul_8.0.1AvailablerheMac3Oct. 2010Beijing Genomics Institute CR_1.0AvailablerheMac2Jan. 2006Baylor College of Medicine HGSC v1.0 Mmul_051212AvailablerheMac1Jan. 2005Baylor College of Medicine HGSC Mmul_0.1ArchivedRock hyraxproCap1Jul. 2008Baylor College of Medicine HGSC Procap1.0AvailableSheepoviAri4Dec. 2015ISGC Oar_v4.0AvailableoviAri3Aug. 2012ISGC Oar_v3.1AvailableoviAri1Feb. 2010ISGC Ovis aries 1.0AvailableShrewsorAra2Aug. 2008Broad Institute SorAra2.0AvailablesorAra1Jun. 2006Broad Institute SorAra1.0AvailableSlothchoHof1Jul. 2008Broad Institute ChoHof1.0AvailableSquirrelspeTri2Nov. 2011Broad Institute SpeTri2.0AvailableSquirrel monkeysaiBol1Oct. 2011Broad Institute SaiBol1.0AvailableTarsiertarSyr2Sep. 2013WashU Tarsius_syrichta-2.0.1AvailabletarSyr1Aug. 2008WUSTL/Broad Institute Tarsyr1.0AvailableTasmanian devilsarHar1Feb. 2011Wellcome Trust Sanger Institute Devil_refv7.0AvailableTenrecechTel2Nov. 2012Broad Institute EchTel2.0AvailableechTel1Jul. 2005Broad Institute echTel1AvailableTree shrewtupBel1Dec. 2006Broad Institute Tupbel1.0AvailableWallabymacEug2Sep. 2009Tammar Wallaby Genome Sequencing Consortium Meug_1.1AvailableWhite rhinoceroscerSim1May 2012Broad Institute CerSimSim1.0AvailableVERTEBRATESAfrican clawed frogxenLae2Aug. 2016Int. Xenopus Sequencing ConsortiumAvailableAmerican alligatorallMis1Aug. 2012Int. Crocodilian Genomes Working Group allMis0.2AvailableAtlantic codgadMor1May 2010Genofisk GadMor_May2010AvailableBudgerigarmelUnd1Sep. 2011WUSTL v6.3AvailableChickengalGal6Mar. 2018GRCg6 Gallus-gallus-6.0AvailablegalGal5Dec. 2015ICGC Gallus-gallus-5.0AvailablegalGal4Nov. 2011ICGC Gallus-gallus-4.0AvailablegalGal3May 2006WUSTL Gallus-gallus-2.1AvailablegalGal2Feb. 2004WUSTL Gallus-gallus-1.0AvailableCoelacanthlatCha1Aug. 2011Broad Institute LatCha1AvailableElephant sharkcalMil1Dec. 2013IMCB Callorhinchus_milli_6.1.3AvailableFugufr3Oct. 2011JGI v5.0Availablefr2Oct. 2004JGI v4.0Availablefr1Aug. 2002JGI v3.0AvailableLampreypetMar3Dec. 2017University of Kentucky Pmar_germline 1.0AvailablepetMar2Sep. 2010WUGSC 7.0AvailablepetMar1Mar. 2007WUSTL v3.0AvailableLizardanoCar2May 2010Broad Institute AnoCar2AvailableanoCar1Feb. 2007Broad Institute AnoCar1AvailableMedakaoryLat2Oct. 2005NIG v1.0AvailableMedium ground finchgeoFor1Apr. 2012BGI GeoFor_1.0 / NCBI 13302AvailableNile tilapiaoreNil2Jan. 2011Broad Institute Release OreNil1.1AvailablePainted turtlechrPic1Dec. 2011IPTGSC Chrysemys_picta_bellii-3.0.1AvailableSticklebackgasAcu1Feb. 2006Broad Institute Release 1.0AvailableTetraodontetNig2Mar. 2007Genoscope v7AvailabletetNig1Feb. 2004Genoscope v7AvailableTibetan frognanPar1Mar. 2015Beijing Genomics Institute BGI_ZX_20015AvailableTurkeymelGal5Nov. 2014Turkey Genome Consortium v5.0AvailablemelGal1Dec. 2009Turkey Genome Consortium v2.01AvailableX. tropicalisxenTro9Jul. 2016JGI v.9.1AvailablexenTro7Sep. 2012JGI v.7.0AvailablexenTro3Nov. 2009JGI v.4.2AvailablexenTro2Aug. 2005JGI v.4.1AvailablexenTro1Oct. 2004JGI v.3.0AvailableZebra finchtaeGut2Feb. 2013WashU taeGut324AvailabletaeGut1Jul. 2008WUSTL v3.2.4AvailableZebrafishdanRer11May 2017Genome Reference Consortium GRCz11AvailabledanRer10Sep. 2014Genome Reference Consortium GRCz10AvailabledanRer7Jul. 2010Sanger Institute Zv9AvailabledanRer6Dec. 2008Sanger Institute Zv8AvailabledanRer5Jul. 2007Sanger Institute Zv7AvailabledanRer4Mar. 2006Sanger Institute Zv6AvailabledanRer3May 2005Sanger Institute Zv5AvailabledanRer2Jun. 2004Sanger Institute Zv4ArchiveddanRer1Nov. 2003Sanger Institute Zv3ArchivedDEUTEROSTOMESC. intestinalisci3Apr. 2011Kyoto KHAvailableC. intestinalisci2Mar. 2005JGI v2.0Availableci1Dec. 2002JGI v1.0AvailableLanceletbraFlo1Mar. 2006JGI v1.0AvailableS. purpuratusstrPur2Sep. 2006Baylor College of Medicine HGSC v. Spur 2.1AvailablestrPur1Apr. 2005Baylor College of Medicine HGSC v. Spur_0.5AvailableINSECTSA. melliferaapiMel2Jan. 2005Baylor College of Medicine HGSC v.Amel_2.0AvailableapiMel1Jul. 2004Baylor College of Medicine HGSC v.Amel_1.2AvailableA. gambiaeanoGam3Oct. 2006International Consortium for the Sequencing of Anopheles Genome AgamP3AvailableanoGam1Feb. 2003IAGP v.MOZ2AvailableD. ananassaedroAna2Aug. 2005Agencourt Arachne releaseAvailabledroAna1Jul. 2004TIGR Celera releaseAvailableD. erectadroEre1Aug. 2005Agencourt Arachne releaseAvailableD. grimshawidroGri1Aug. 2005Agencourt Arachne releaseAvailableD. melanogasterdm6Aug. 2014BDGP Release 6 + ISO1 MTAvailabledm3Apr. 2006BDGP Release 5Availabledm2Apr. 2004BDGP Release 4Availabledm1Jan. 2003BDGP Release 3AvailableD. mojavensisdroMoj2Aug. 2005Agencourt Arachne releaseAvailabledroMoj1Aug. 2004Agencourt Arachne releaseAvailableD. persimilisdroPer1Oct. 2005Broad Institute releaseAvailableD. pseudoobscuradp3Nov. 2004FlyBase Release 1.0Availabledp2Aug. 2003Baylor College of Medicine HGSC Freeze 1AvailableD. sechelliadroSec1Oct. 2005Broad Institute Release 1.0AvailableD. simulansdroSim1Apr. 2005WUSTL Release 1.0AvailableD. virilisdroVir2Aug. 2005Agencourt Arachne releaseAvailabledroVir1Jul. 2004Agencourt Arachne releaseAvailableD. yakubadroYak2Nov. 2005WUSTL Release 2.0AvailabledroYak1Apr. 2004WUSTL Release 1.0AvailableNEMATODESC. brennericaePb2Feb. 2008WUSTL 6.0.1AvailablecaePb1Jan. 2007WUSTL 4.0AvailableC. briggsaecb3Jan. 2007WUSTL Cb3Availablecb1Jul. 2002WormBase v. cb25.agp8AvailableC. elegansce11Feb. 2013C. elegans Sequencing Consortium WBcel235Availablece10Oct. 2010WormBase v. WS220Availablece6May 2008WormBase v. WS190Availablece4Jan. 2007WormBase v. WS170Availablece2Mar. 2004WormBase v. WS120Availablece1May 2003WormBase v. WS100ArchivedC. japonicacaeJap1Mar. 2008WUSTL 3.0.2AvailableC. remaneicaeRem3May 2007WUSTL 15.0.1AvailablecaeRem2Mar. 2006WUSTL 1.0AvailableP. pacificuspriPac1Feb. 2007WUSTL 5.0AvailableOTHERSea HareaplCal1Sep. 2008Broad Release Aplcal2.0AvailableYeastsacCer3April 2011SGD April 2011 sequenceAvailablesacCer2June 2008SGD June 2008 sequenceAvailablesacCer1Oct. 2003SGD 1 Oct 2003 sequenceAvailableVIRUSESEbola ViruseboVir3June 2014Sierra Leone 2014 (G3683/KM034562.1)AvailableSARS-CoV-2wuhCor1Jan. 2020SARS-CoV-2 ASM985889v3Available
https://www.ncbi.nlm.nih.gov/grc
http://genomeref.blogspot.com/
| 
 随着时间的推移,在更先进技术的加持下,在已经构建好的基因组和注释信息上不断增加,删减,修改,就有了不同的版本。而每一个版本的参考基因组都会对应有一个基因组注释文件(天书和字典一一对应),接下来我们看看参考基因组版本是怎么指定的。
随着时间的推移,在更先进技术的加持下,在已经构建好的基因组和注释信息上不断增加,删减,修改,就有了不同的版本。而每一个版本的参考基因组都会对应有一个基因组注释文件(天书和字典一一对应),接下来我们看看参考基因组版本是怎么指定的。








